Genetic Alternatives for Experimental Adaptation to Colistin in Three Pseudomonas aeruginosa Lineages
- PMID: 38786180
- PMCID: PMC11117860
- DOI: 10.3390/antibiotics13050452
Genetic Alternatives for Experimental Adaptation to Colistin in Three Pseudomonas aeruginosa Lineages
Abstract
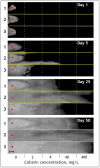
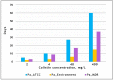
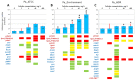
Pseudomonas aeruginosa is characterized by a high adaptive potential, developing resistance in response to antimicrobial pressure. We employed a spatiotemporal evolution model to disclose the pathways of adaptation to colistin, a last-resort polymyxin antimicrobial, among three unrelated P. aeruginosa lineages. The P. aeruginosa ATCC-27833 reference strain (Pa_ATCC), an environmental P. aeruginosa isolate (Pa_Environment), and a clinical isolate with multiple drug resistance (Pa_MDR) were grown over an increasing 5-step colistin concentration gradient from 0 to 400 mg/L. Pa_Environment demonstrated the highest growth pace, achieving the 400 mg/L band in 15 days, whereas it took 37 and 60 days for Pa_MDR and Pa_ATCC, respectively. To identify the genome changes that occurred during adaptation to colistin, the isolates selected during the growth of the bacteria (n = 185) were subjected to whole genome sequencing. In total, 17 mutation variants in eight lipopolysaccharide-synthesis-associated genes were detected. phoQ and lpxL/PA0011 were affected in all three lineages, whereas changes in pmrB were found in Pa_Environment and Pa_MDR but not in Pa_ATCC. In addition, mutations were detected in 34 general metabolism genes, and each lineage developed mutations in a unique set of such genes. Thus, the three examined distinct P. aeruginosa strains demonstrated different capabilities and genetic pathways of colistin adaptation.
Keywords: Pseudomonas aeruginosa; antibiotic resistance; colistin; experimental evolution; mutation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
The PitA protein contributes to colistin susceptibility in Pseudomonas aeruginosa.PLoS One. 2023 Oct 12;18(10):e0292818. doi: 10.1371/journal.pone.0292818. eCollection 2023. PLoS One. 2023. PMID: 37824582 Free PMC article.
-
Detection of Colistin Resistance in Pseudomonas aeruginosa Using the MALDIxin Test on the Routine MALDI Biotyper Sirius Mass Spectrometer.Front Microbiol. 2021 Aug 31;12:725383. doi: 10.3389/fmicb.2021.725383. eCollection 2021. Front Microbiol. 2021. PMID: 34531843 Free PMC article.
-
PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients.Antimicrob Agents Chemother. 2012 Feb;56(2):1019-30. doi: 10.1128/AAC.05829-11. Epub 2011 Nov 21. Antimicrob Agents Chemother. 2012. PMID: 22106224 Free PMC article.
-
A Review of Resistance to Polymyxins and Evolving Mobile Colistin Resistance Gene (mcr) among Pathogens of Clinical Significance.Antibiotics (Basel). 2023 Nov 6;12(11):1597. doi: 10.3390/antibiotics12111597. Antibiotics (Basel). 2023. PMID: 37998799 Free PMC article. Review.
-
Colistin in multi-drug resistant Pseudomonas aeruginosa blood-stream infections: a narrative review for the clinician.J Infect. 2014 Jul;69(1):1-12. doi: 10.1016/j.jinf.2014.03.001. Epub 2014 Mar 11. J Infect. 2014. PMID: 24631777 Review.
Cited by
-
Deciphering Multidrug-Resistant Pseudomonas aeruginosa: Mechanistic Insights and Environmental Risks.Toxics. 2025 Apr 12;13(4):303. doi: 10.3390/toxics13040303. Toxics. 2025. PMID: 40278619 Free PMC article. Review.
-
In Vitro Resistance-Predicting Studies and In Vitro Resistance-Related Parameters-A Hit-to-Lead Perspective.Pharmaceuticals (Basel). 2024 Aug 15;17(8):1068. doi: 10.3390/ph17081068. Pharmaceuticals (Basel). 2024. PMID: 39204172 Free PMC article. Review.
-
High-risk Pseudomonas aeruginosa clones harboring β-lactamases: 2024 update.Heliyon. 2024 Dec 26;11(1):e41540. doi: 10.1016/j.heliyon.2024.e41540. eCollection 2025 Jan 15. Heliyon. 2024. PMID: 39850428 Free PMC article.
References
-
- Labovska S. Pseudomonas aeruginosa as a Cause of Nosocomial Infections. In: Das T., editor. Pseudomonas aeruginosa—Biofilm Formation, Infections and Treatments. IntechOpen; London, UK: 2021. - DOI
-
- Wheatley R., Diaz Caballero J., Kapel N., De Winter F.H., Jangir P., Quinn A., del Barrio-Tofiño E., López-Causapé C., Hedge J., Torrens G., et al. Rapid evolution and host immunity drive the rise and fall of carbapenem resistance during an acute Pseudomonas aeruginosa infection. Nat. Commun. 2021;12:2460. doi: 10.1038/s41467-021-22814-9. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

