Strategies for the Enhancement of Secondary Metabolite Production via Biosynthesis Gene Cluster Regulation in Aspergillus oryzae
- PMID: 38786667
- PMCID: PMC11121810
- DOI: 10.3390/jof10050312
Strategies for the Enhancement of Secondary Metabolite Production via Biosynthesis Gene Cluster Regulation in Aspergillus oryzae
Abstract
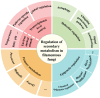
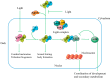
The filamentous fungus Aspergillus oryzae (A. oryzae) has been extensively used for the biosynthesis of numerous secondary metabolites with significant applications in agriculture and food and medical industries, among others. However, the identification and functional prediction of metabolites through genome mining in A. oryzae are hindered by the complex regulatory mechanisms of secondary metabolite biosynthesis and the inactivity of most of the biosynthetic gene clusters involved. The global regulatory factors, pathway-specific regulatory factors, epigenetics, and environmental signals significantly impact the production of secondary metabolites, indicating that appropriate gene-level modulations are expected to promote the biosynthesis of secondary metabolites in A. oryzae. This review mainly focuses on illuminating the molecular regulatory mechanisms for the activation of potentially unexpressed pathways, possibly revealing the effects of transcriptional, epigenetic, and environmental signal regulation. By gaining a comprehensive understanding of the regulatory mechanisms of secondary metabolite biosynthesis, strategies can be developed to enhance the production and utilization of these metabolites, and potential functions can be fully exploited.
Keywords: Aspergillus oryzae; regulatory gene; secondary metabolism; secondary metabolites.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Sequencing and functional annotation of the whole genome of the filamentous fungus Aspergillus westerdijkiae.BMC Genomics. 2016 Aug 15;17(1):633. doi: 10.1186/s12864-016-2974-x. BMC Genomics. 2016. PMID: 27527502 Free PMC article.
-
Use of the kojA promoter, involved in kojic acid biosynthesis, for polyketide production in Aspergillus oryzae: implications for long-term production.BMC Biotechnol. 2019 Oct 26;19(1):70. doi: 10.1186/s12896-019-0567-x. BMC Biotechnol. 2019. PMID: 31655589 Free PMC article.
-
Beyond the Biosynthetic Gene Cluster Paradigm: Genome-Wide Coexpression Networks Connect Clustered and Unclustered Transcription Factors to Secondary Metabolic Pathways.Microbiol Spectr. 2021 Oct 31;9(2):e0089821. doi: 10.1128/Spectrum.00898-21. Epub 2021 Sep 15. Microbiol Spectr. 2021. PMID: 34523946 Free PMC article.
-
Biosynthesis and biological function of secondary metabolites of the rice blast fungus Pyricularia oryzae.J Ind Microbiol Biotechnol. 2021 Dec 23;48(9-10):kuab058. doi: 10.1093/jimb/kuab058. J Ind Microbiol Biotechnol. 2021. PMID: 34379774 Free PMC article. Review.
-
Molecular regulation of fungal secondary metabolism.World J Microbiol Biotechnol. 2023 May 20;39(8):204. doi: 10.1007/s11274-023-03649-6. World J Microbiol Biotechnol. 2023. PMID: 37209190 Review.
Cited by
-
Synthetic biology meets Aspergillus: engineering strategies for next-generation organic acid production.World J Microbiol Biotechnol. 2025 Jan 13;41(2):36. doi: 10.1007/s11274-024-04246-x. World J Microbiol Biotechnol. 2025. PMID: 39800796 Review.
-
Involvement of LaeA and Velvet Proteins in Regulating the Production of Mycotoxins and Other Fungal Secondary Metabolites.J Fungi (Basel). 2024 Aug 8;10(8):561. doi: 10.3390/jof10080561. J Fungi (Basel). 2024. PMID: 39194887 Free PMC article. Review.
-
A Close View of the Production of Bioactive Fungal Metabolites Mediated by Chromatin Modifiers.Molecules. 2024 Jul 27;29(15):3536. doi: 10.3390/molecules29153536. Molecules. 2024. PMID: 39124942 Free PMC article. Review.
-
Light-Regulated Gene Expression Patterns During Conidial Formation in Aspergillus oryzae.Curr Issues Mol Biol. 2025 May 20;47(5):373. doi: 10.3390/cimb47050373. Curr Issues Mol Biol. 2025. PMID: 40699772 Free PMC article.
-
Construction of an Efficient Engineered Strain for Chaetoglobosin A Bioresource Production from Potato Starch Industrial Waste.Foods. 2025 Feb 28;14(5):842. doi: 10.3390/foods14050842. Foods. 2025. PMID: 40077545 Free PMC article.
References
Publication types
Grants and funding
- 32200606/National Natural Science Foundation of China
- 2021YFA1301302/Key Special Projects of the National Key Research and Development Plan
- 20213AAG02020/Project of the Department of Science and Technology of the Jiangxi Province
- JSZZ202301021/Self-made Experimental Instruments and Equipment Project of Shenzhen Technology University
LinkOut - more resources
Full Text Sources

