The Putative Cytochrome b 5 Domain-Containing Protein CaDap1 Homologue Is Involved in Antifungal Drug Tolerance, Cell Wall Chitin Maintenance, and Virulence in Candida albicans
- PMID: 38786671
- PMCID: PMC11122062
- DOI: 10.3390/jof10050316
The Putative Cytochrome b 5 Domain-Containing Protein CaDap1 Homologue Is Involved in Antifungal Drug Tolerance, Cell Wall Chitin Maintenance, and Virulence in Candida albicans
Abstract
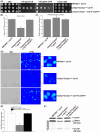
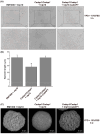
Candida albicans (Ca), a prominent opportunistic fungal pathogen in humans, has garnered considerable attention due to its infectious properties. Herein, we have identified and characterized CaCDAP1 (Ca orf19.1034), a homolog of ScDAP1 found in Saccharomyces cerevisiae. CaCDAP1 encodes a 183-amino acid protein with a conserved cytochrome b5-like heme-binding domain. The deletion of CaDAP1 renders Ca cells susceptible to caspofungin and terbinafine. CaDAP1 deletion confers resistance to Congo Red and Calcofluor White, and sensitivity to sodium dodecyl sulfate. The deletion of CaDAP1 results in a 50% reduction in chitin content within the cell wall, the downregulation of phosphorylation levels in CaMkc1, and the upregulation of phosphorylation levels in CaCek1. Notably, CaDAP1 deletion results in the abnormal hyphal development of Ca cells and diminishes virulence in a mouse systemic infection model. Thus, CaDAP1 emerges as a critical regulator governing cellular responses to antifungal drugs, the synthesis of cell wall chitin, and virulence in Ca.
Keywords: CaDAP1; Candida albicans; chitin content; drug susceptibility; virulence.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Genotypic and phenotypic characterization of Candida albicans Lebanese hospital isolates resistant and sensitive to caspofungin.Fungal Genet Biol. 2019 Jun;127:12-22. doi: 10.1016/j.fgb.2019.02.008. Epub 2019 Feb 20. Fungal Genet Biol. 2019. PMID: 30794951
-
The lipid flippase subunit Cdc50 is required for antifungal drug resistance, endocytosis, hyphal development and virulence in Candida albicans.FEMS Yeast Res. 2019 May 1;19(3):foz033. doi: 10.1093/femsyr/foz033. FEMS Yeast Res. 2019. PMID: 31004489
-
The CRH family coding for cell wall glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans.J Biol Chem. 2006 Dec 29;281(52):40399-411. doi: 10.1074/jbc.M606361200. Epub 2006 Oct 30. J Biol Chem. 2006. PMID: 17074760
-
Candida and candidaemia. Susceptibility and epidemiology.Dan Med J. 2013 Nov;60(11):B4698. Dan Med J. 2013. PMID: 24192246 Review.
-
Cell wall associated proteins involved in filamentation with impact on the virulence of Candida albicans.Microbiol Res. 2022 May;258:126996. doi: 10.1016/j.micres.2022.126996. Epub 2022 Feb 22. Microbiol Res. 2022. PMID: 35247799 Review.
Cited by
-
The Mediator complex subunit MoMed15 plays an important role in conferring sensitivity to isoprothiolane by modulating xenobiotic metabolism in M. oryzae.mBio. 2024 Dec 11;15(12):e0177824. doi: 10.1128/mbio.01778-24. Epub 2024 Nov 12. mBio. 2024. PMID: 39530687 Free PMC article.
References
Grants and funding
- 1808085MC74/the Anhui Provincial Natural Science Foundation
- KJ2018A0676/the University Natural Science Research Key projects in Anhui Province
- gxgnfx2021118/the Anhui Province University Outstanding Talents Cultivation Funding Project
- 2023AH010045/the Anhui Province University Innovation Team Project
LinkOut - more resources
Full Text Sources

