Hepatic Activin E mediates liver-adipose inter-organ communication, suppressing adipose lipolysis in response to elevated serum fatty acids
- PMID: 38787338
- PMCID: PMC10656223
- DOI: 10.1016/j.molmet.2023.101830
Hepatic Activin E mediates liver-adipose inter-organ communication, suppressing adipose lipolysis in response to elevated serum fatty acids
Abstract
Objective: The liver is a central regulator of energy metabolism exerting its influence both through intrinsic processing of substrates such as glucose and fatty acid as well as by secreting endocrine factors, known as hepatokines, which influence metabolism in peripheral tissues. Human genome wide association studies indicate that a predicted loss-of-function variant in the Inhibin βE gene (INHBE), encoding the putative hepatokine Activin E, is associated with reduced abdominal fat mass and cardiometabolic disease risk. However, the regulation of hepatic Activin E and the influence of Activin E on adiposity and metabolic disease are not well understood. Here, we examine the relationship between hepatic Activin E and adipose metabolism, testing the hypothesis that Activin E functions as part of a liver-adipose, inter-organ feedback loop to suppress adipose tissue lipolysis in response to elevated serum fatty acids and hepatic fatty acid exposure.
Methods: The relationship between hepatic Activin E and non-esterified fatty acids (NEFA) released from adipose lipolysis was assessed in vivo using fasted CL 316,243 treated mice and in vitro using Huh7 hepatocytes treated with fatty acids. The influence of Activin E on adipose lipolysis was examined using a combination of Inhbe knockout mice, a mouse model of hepatocyte-specific overexpression of Activin E, and mouse brown adipocytes treated with Activin E enriched media.
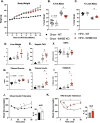
Results: Increasing hepatocyte NEFA exposure in vivo by inducing adipose lipolysis through fasting or CL 316,243 treatment increased hepatic Inhbe expression. Similarly, incubation of Huh7 human hepatocytes with fatty acids increased expression of INHBE. Genetic ablation of Inhbe in mice increased fasting circulating NEFA and hepatic triglyceride accumulation. Treatment of mouse brown adipocytes with Activin E conditioned media and overexpression of Activin E in mice suppressed adipose lipolysis and reduced serum FFA levels, respectively. The suppressive effects of Activin E on lipolysis were lost in CRISPR-mediated ALK7 deficient cells and ALK7 kinase deficient mice. Disruption of the Activin E-ALK7 signaling axis in Inhbe KO mice reduced adiposity upon HFD feeding, but caused hepatic steatosis and insulin resistance.
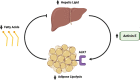
Conclusions: Taken together, our data suggest that Activin E functions as part of a liver-adipose feedback loop, such that in response to increased serum free fatty acids and elevated hepatic triglyceride, Activin E is released from hepatocytes and signals in adipose through ALK7 to suppress lipolysis, thereby reducing free fatty acid efflux to the liver and preventing excessive hepatic lipid accumulation. We find that disrupting this Activin E-ALK7 inter-organ communication network by ablation of Inhbe in mice increases lipolysis and reduces adiposity, but results in elevated hepatic triglyceride and impaired insulin sensitivity. These results highlight the liver-adipose, Activin E-ALK7 signaling axis as a critical regulator of metabolic homeostasis.
Keywords: Adipose tissue; Diabetes; Lipolysis; Obesity.
Copyright © 2023 The Authors. Published by Elsevier GmbH.. All rights reserved.
Figures




References
-
- Sugiyama M., Kikuchi A., Misu H., Igawa H., Ashihara M., Kushima Y., et al. Inhibin βE (INHBE) is a possible insulin resistance-associated hepatokine identified by comprehensive gene expression analysis in human liver biopsy samples. PLoS One. 2018;13(3) doi: 10.1371/journal.pone.0194798. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

