SLAMF8 can predict prognosis of pan-cancer and the immunotherapy response effectivity of gastric cancer
- PMID: 38787377
- PMCID: PMC11164479
- DOI: 10.18632/aging.205850
SLAMF8 can predict prognosis of pan-cancer and the immunotherapy response effectivity of gastric cancer
Abstract
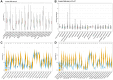
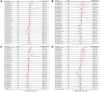
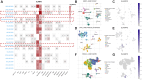
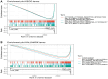
SLAMF8, the eighth member of the Signaling Lymphocytic Activation Molecule Family (SLAMF), functions in the regulation of the development and activity of diverse immune cells as a costimulatory receptor within the SLAMF family. Studies had revealed that SLAMF8 is expressed higher in several autoimmune inflammation diseases and tumors. Nevertheless, the connection between SLAMF8 and pan-cancer remains undisclosed. The research investigated the correlation between SLAMF8 and various factors including the immune microenvironment, microsatellite instability, immune novel antigen, gene mutation, immune regulatory factors, immune blockade TMB, and immune or molecular subtypes of SLAMF8 in verse cancer types. Immunohistochemistry was ultimately employed to validate the presence of the SLAMF8 gene in various tumor types including hepatocellular carcinoma, prostate adenocarcinoma, and kidney renal clear cell carcinoma. Furthermore, the relationship between SLAMF8 expression and the therapeutic efficacy of the PD1 blockade agent, Sintilimab, treatment in gastric cancer was validated. The result of differential analysis suggested that SLAMF8 was over-expressed in pan-cancer compared with paracancerous tissues. The analysis of survival indicated a connection between SLAMF8 and the overall prognosis in different types of cancers, where higher levels of SLAMF8 were found to be significantly linked to unfavorable outcomes in patients but favorable outcome of immunotherapy in gastric cancer. Significant correlations were observed between SLAMF8 levels and pan-cancer tumorigenesis, tumor metabolism, and immunity. As a result, SLAMF8 may become an important prognostic biomarker in the majority of tumors and a hopeful gene target for immunotherapy against gastric cancer.
Keywords: SLAMF8; TME; biomarker; immunotherapy; pan-cancer.
Conflict of interest statement
Figures









Similar articles
-
Costimulatory checkpoint SLAMF8 is an independent prognosis factor in glioma.CNS Neurosci Ther. 2019 Mar;25(3):333-342. doi: 10.1111/cns.13041. Epub 2018 Aug 13. CNS Neurosci Ther. 2019. PMID: 30105842 Free PMC article.
-
Pan-cancer and experimental analyses reveal the immunotherapeutic significance of CST2 and its association with stomach adenocarcinoma proliferation and metastasis.Front Immunol. 2025 Jan 24;15:1466806. doi: 10.3389/fimmu.2024.1466806. eCollection 2024. Front Immunol. 2025. PMID: 39926600 Free PMC article.
-
UBR1 is a prognostic biomarker and therapeutic target associated with immune cell infiltration in gastric cancer.Aging (Albany NY). 2024 Aug 23;16(16):12029-12049. doi: 10.18632/aging.206079. Epub 2024 Aug 23. Aging (Albany NY). 2024. PMID: 39181686 Free PMC article.
-
SLAMF8 (BLAME) as a novel immune checkpoint: Implications for inflammation, autoimmunity, and oncology.Pathol Res Pract. 2025 Aug;272:156072. doi: 10.1016/j.prp.2025.156072. Epub 2025 Jun 3. Pathol Res Pract. 2025. PMID: 40513428 Review.
-
Signaling lymphocytic activation molecule family receptors as potential immune therapeutic targets in solid tumors.Front Immunol. 2024 Feb 27;15:1297473. doi: 10.3389/fimmu.2024.1297473. eCollection 2024. Front Immunol. 2024. PMID: 38476238 Free PMC article. Review.
Cited by
-
Spatial transcriptomic analysis of 4NQO-induced tongue cancer revealed cellular lineage diversity and evolutionary trajectory.Front Oncol. 2025 Jul 3;15:1592044. doi: 10.3389/fonc.2025.1592044. eCollection 2025. Front Oncol. 2025. PMID: 40678065 Free PMC article.
-
DNA methylation expression patterns predict outcome of clear cell renal cell carcinoma.Discov Oncol. 2025 May 26;16(1):934. doi: 10.1007/s12672-025-02764-0. Discov Oncol. 2025. PMID: 40419839 Free PMC article.
-
SLAMF receptors: key regulators of tumor progression and emerging targets for cancer immunotherapy.Mol Cancer. 2025 May 17;24(1):145. doi: 10.1186/s12943-025-02308-8. Mol Cancer. 2025. PMID: 40382610 Free PMC article. Review.
References
-
- de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J, Ji SG, Heap G, Nimmo ER, Edwards C, et al.. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017; 49:256–61. 10.1038/ng.3760 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

