Secreted dengue virus NS1 from infection is predominantly dimeric and in complex with high-density lipoprotein
- PMID: 38787378
- PMCID: PMC11126310
- DOI: 10.7554/eLife.90762
Secreted dengue virus NS1 from infection is predominantly dimeric and in complex with high-density lipoprotein
Abstract
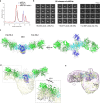
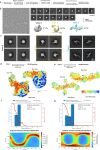
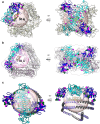
Severe dengue infections are characterized by endothelial dysfunction shown to be associated with the secreted nonstructural protein 1 (sNS1), making it an attractive vaccine antigen and biotherapeutic target. To uncover the biologically relevant structure of sNS1, we obtained infection-derived sNS1 (isNS1) from dengue virus (DENV)-infected Vero cells through immunoaffinity purification instead of recombinant sNS1 (rsNS1) overexpressed in insect or mammalian cell lines. We found that isNS1 appeared as an approximately 250 kDa complex of NS1 and ApoA1 and further determined the cryoEM structures of isNS1 and its complex with a monoclonal antibody/Fab. Indeed, we found that the major species of isNS1 is a complex of the NS1 dimer partially embedded in a high-density lipoprotein (HDL) particle. Crosslinking mass spectrometry studies confirmed that the isNS1 interacts with the major HDL component ApoA1 through interactions that map to the NS1 wing and hydrophobic domains. Furthermore, our studies demonstrated that the sNS1 in sera from DENV-infected mice and a human patient form a similar complex as isNS1. Our results report the molecular architecture of a biological form of sNS1, which may have implications for the molecular pathogenesis of dengue.
Keywords: biomarker; cryoEM; dengue infection; flavivirus; high-density lipoprotein; human; infectious disease; microbiology; molecular biophysics; mouse; pathogenesis factor; structural biology.
© 2023, Chew, Ngoh, Phoo et al.
Conflict of interest statement
BC, AN, WP, KC, ZS, NT, SL, MW, SW, MC, JL, EO, CR, RS, SV, DL No competing interests declared
Figures















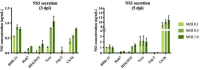

Update of
- doi: 10.1101/2022.04.06.487425
- doi: 10.7554/eLife.90762.1
- doi: 10.7554/eLife.90762.2
References
-
- Alcalá AC, Hernández-Bravo R, Medina F, Coll DS, Zambrano JL, Del Angel RM, Ludert JE. The dengue virus non-structural protein 1 (NS1) is secreted from infected mosquito cells via a non-classical caveolin-1-dependent pathway. The Journal of General Virology. 2017;98:2088–2099. doi: 10.1099/jgv.0.000881. - DOI - PubMed
-
- Alcon S, Talarmin A, Debruyne M, Falconar A, Deubel V, Flamand M. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. Journal of Clinical Microbiology. 2002;40:376–381. doi: 10.1128/JCM.40.02.376-381.2002. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- T2EP30220-0020/Ministry of Education - Singapore
- MOH-OFIRG20nov-0002/National Medical Research Council
- LKCMedicine Dean's Postdoctoral Fellowship/Lee Kong Chian School of Medicine, Nanyang Technological University
- MOHOFIRG18may-0006/National Medical Research Council
- Career Development Fund 2021/Agency for Science, Technology and Research
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

