Conifers Concentrate Large Numbers of NLR Immune Receptor Genes on One Chromosome
- PMID: 38787537
- PMCID: PMC11171428
- DOI: 10.1093/gbe/evae113
Conifers Concentrate Large Numbers of NLR Immune Receptor Genes on One Chromosome
Abstract
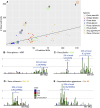
Nucleotide-binding domain and leucine-rich repeat (NLR) immune receptor genes form a major line of defense in plants, acting in both pathogen recognition and resistance machinery activation. NLRs are reported to form large gene clusters in limber pine (Pinus flexilis), but it is unknown how widespread this genomic architecture may be among the extant species of conifers (Pinophyta). We used comparative genomic analyses to assess patterns in the abundance, diversity, and genomic distribution of NLR genes. Chromosome-level whole genome assemblies and high-density linkage maps in the Pinaceae, Cupressaceae, Taxaceae, and other gymnosperms were scanned for NLR genes using existing and customized pipelines. The discovered genes were mapped across chromosomes and linkage groups and analyzed phylogenetically for evolutionary history. Conifer genomes are characterized by dense clusters of NLR genes, highly localized on one chromosome. These clusters are rich in TNL-encoding genes, which seem to have formed through multiple tandem duplication events. In contrast to angiosperms and nonconiferous gymnosperms, genomic clustering of NLR genes is ubiquitous in conifers. NLR-dense genomic regions are likely to influence a large part of the plant's resistance, informing our understanding of adaptation to biotic stress and the development of genetic resources through breeding.
Keywords: NBS-LRR; comparative genomics; gene clusters; gene family evolution; genomic architecture; resistance genes.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest The authors declare that they have no competing interests.
Figures



Similar articles
-
The large repertoire of conifer NLR resistance genes includes drought responsive and highly diversified RNLs.Sci Rep. 2019 Aug 12;9(1):11614. doi: 10.1038/s41598-019-47950-7. Sci Rep. 2019. PMID: 31406137 Free PMC article.
-
Fine dissection of limber pine resistance to Cronartium ribicola using targeted sequencing of the NLR family.BMC Genomics. 2021 Jul 23;22(1):567. doi: 10.1186/s12864-021-07885-8. BMC Genomics. 2021. PMID: 34294045 Free PMC article.
-
Limber pine (Pinus flexilis James) genetic map constructed by exome-seq provides insight into the evolution of disease resistance and a genomic resource for genomics-based breeding.Plant J. 2019 May;98(4):745-758. doi: 10.1111/tpj.14270. Epub 2019 Mar 1. Plant J. 2019. PMID: 30729601
-
Activation and Regulation of NLR Immune Receptor Networks.Plant Cell Physiol. 2022 Oct 31;63(10):1366-1377. doi: 10.1093/pcp/pcac116. Plant Cell Physiol. 2022. PMID: 35941738 Review.
-
NLR receptors in plant immunity: making sense of the alphabet soup.EMBO Rep. 2023 Oct 9;24(10):e57495. doi: 10.15252/embr.202357495. Epub 2023 Aug 21. EMBO Rep. 2023. PMID: 37602936 Free PMC article. Review.
Cited by
-
NRC Immune receptor networks show diversified hierarchical genetic architecture across plant lineages.Plant Cell. 2024 Sep 3;36(9):3399-3418. doi: 10.1093/plcell/koae179. Plant Cell. 2024. PMID: 38922300 Free PMC article.
References
-
- Amerson HV, Nelson C, Kubisiak T, Kuhlman E, Garcia S. Identification of nine pathotype-specific genes conferring resistance to fusiform rust in loblolly pine (Pinus taeda L.). Forests. 2015:6(12):2739–2761. 10.3390/f6082739. - DOI
-
- Avni R, Lux T, Minz-Dub A, Millet E, Sela H, Distelfeld A, Deek J, Yu G, Steuernagel B, Pozniak C, et al. Genome sequences of three Aegilops species of the section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. Plant J. 2022:110(1):179–192. 10.1111/tpj.15664. - DOI - PMC - PubMed
-
- Bedon F, Bomal C, Caron S, Levasseur C, Boyle B, Mansfield SD, Schmidt A, Gershenzon J, Grima-Pettenati J, Séguin A. Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J Exp Bot. 2010:61(14):3847–3864. 10.1093/jxb/erq196. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

