Efficient Segmental Isotope Labeling of Integral Membrane Proteins for High-Resolution NMR Studies
- PMID: 38787792
- PMCID: PMC11157531
- DOI: 10.1021/jacs.4c03294
Efficient Segmental Isotope Labeling of Integral Membrane Proteins for High-Resolution NMR Studies
Abstract
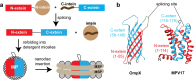
High-resolution structural NMR analyses of membrane proteins are challenging due to their large size, resulting in broad resonances and strong signal overlap. Among the isotope labeling methods that can remedy this situation, segmental isotope labeling is a suitable strategy to simplify NMR spectra and retain high-resolution structural information. However, protein ligation within integral membrane proteins is complicated since the hydrophobic protein fragments are insoluble, and the removal of ligation side-products is elaborate. Here, we show that a stabilized split-intein system can be used for rapid and high-yield protein trans-splicing of integral membrane proteins under denaturing conditions. This setup enables segmental isotope labeling experiments within folded protein domains for NMR studies. We show that high-quality NMR spectra of markedly reduced complexity can be obtained in detergent micelles and lipid nanodiscs. Of note, the nanodisc insertion step specifically selects for the ligated and correctly folded membrane protein and simultaneously removes ligation byproducts. Using this tailored workflow, we show that high-resolution NMR structure determination is strongly facilitated with just two segmentally isotope-labeled membrane protein samples. The presented method will be broadly applicable to structural and dynamical investigations of (membrane-) proteins and their complexes by solution and solid-state NMR but also other structural methods where segmental labeling is beneficial.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Markus M. A.; Dayie K. T.; Matsudaira P.; Wagner G. Effect of deuteration on the amide proton relaxation rates in proteins. Heteronuclear NMR experiments on villin 14T. J. Magn Reson B 1994, 105 (2), 192–195. 10.1006/jmrb.1994.1122. - DOI - PubMed
- Daniilidis M.; Brandl M. J.; Hagn F. The Advanced Properties of Circularized MSP Nanodiscs Facilitate High-resolution NMR Studies of Membrane Proteins. J. Mol. Biol. 2022, 434 (24), 167861 10.1016/j.jmb.2022.167861. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

