Efficient wastewater sample filtration improves the detection of SARS-CoV-2 variants: An extensive analysis based on sequencing parameters
- PMID: 38787865
- PMCID: PMC11125551
- DOI: 10.1371/journal.pone.0304158
Efficient wastewater sample filtration improves the detection of SARS-CoV-2 variants: An extensive analysis based on sequencing parameters
Abstract
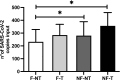
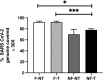
During the SARS-CoV-2 pandemic, many countries established wastewater (WW) surveillance to objectively monitor the level of infection within the population. As new variants continue to emerge, it has become clear that WW surveillance is an essential tool for the early detection of variants. The EU Commission published a recommendation suggesting an approach to establish surveillance of SARS-CoV-2 and its variants in WW, besides specifying the methodology for WW concentration and RNA extraction. Therefore, different groups have approached the issue with different strategies, mainly focusing on WW concentration methods, but only a few groups highlighted the importance of prefiltering WW samples and/or purification of RNA samples. Aiming to obtain high-quality sequencing data allowing variants detection, we compared four experimental conditions generated from the treatment of: i) WW samples by WW filtration and ii) the extracted RNA by DNase treatment, purification and concentration of the extracted RNA. To evaluate the best condition, the results were assessed by focusing on several sequencing parameters, as the outcome of SARS-CoV-2 sequencing from WW is crucial for variant detection. Overall, the best sequencing result was obtained by filtering the WW sample. Moreover, the present study provides an overview of some sequencing parameters to consider when optimizing a method for monitoring SARS-CoV-2 variants from WW samples, which can also be applied to any sample preparation methodology.
Copyright: © 2024 Robotto et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

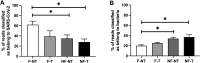



References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

