Longitudinal plasma proteomic analysis of 1117 hospitalized patients with COVID-19 identifies features associated with severity and outcomes
- PMID: 38787940
- PMCID: PMC11122669
- DOI: 10.1126/sciadv.adl5762
Longitudinal plasma proteomic analysis of 1117 hospitalized patients with COVID-19 identifies features associated with severity and outcomes
Abstract
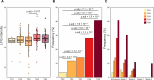
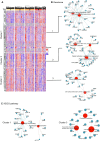
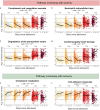
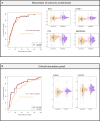
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is characterized by highly heterogeneous manifestations ranging from asymptomatic cases to death for still incompletely understood reasons. As part of the IMmunoPhenotyping Assessment in a COVID-19 Cohort study, we mapped the plasma proteomes of 1117 hospitalized patients with COVID-19 from 15 hospitals across the United States. Up to six samples were collected within ~28 days of hospitalization resulting in one of the largest COVID-19 plasma proteomics cohorts with 2934 samples. Using perchloric acid to deplete the most abundant plasma proteins allowed for detecting 2910 proteins. Our findings show that increased levels of neutrophil extracellular trap and heart damage markers are associated with fatal outcomes. Our analysis also identified prognostic biomarkers for worsening severity and death. Our comprehensive longitudinal plasma proteomics study, involving 1117 participants and 2934 samples, allowed for testing the generalizability of the findings of many previous COVID-19 plasma proteomics studies using much smaller cohorts.
Figures


References
-
- Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L., Ge W., Liu W., Liang S., Chen H., Zhang Y., Li J., Xu J., He Z., Chen B., Wang J., Yan H., Zheng Y., Wang D., Zhu J., Kong Z., Kang Z., Liang X., Ding X., Ruan G., Xiang N., Cai X., Gao H., Li L., Li S., Xiao Q., Lu T., Zhu Y., Liu H., Chen H., Guo T., Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 182, 59–72.e15 (2020). - PMC - PubMed
-
- Wang Z., Cryar A., Lemke O., Tober-Lau P., Ludwig D., Helbig E. T., Hippenstiel S., Sander L. E., Blake D., Lane C. S., Sayers R. L., Mueller C., Zeiser J., Townsend S., Demichev V., Mulleder M., Kurth F., Sirka E., Hartl J., Ralser M., A multiplex protein panel assay for severity prediction and outcome prognosis in patients with COVID-19: An observational multi-cohort study. EClinicalMedicine 49, 101495 (2022). - PMC - PubMed
-
- Demichev V., Tober-Lau P., Lemke O., Nazarenko T., Thibeault C., Whitwell H., Rohl A., Freiwald A., Szyrwiel L., Ludwig D., Correia-Melo C., Aulakh S. K., Helbig E. T., Stubbemann P., Lippert L. J., Gruning N. M., Blyuss O., Vernardis S., White M., Messner C. B., Joannidis M., Sonnweber T., Klein S. J., Pizzini A., Wohlfarter Y., Sahanic S., Hilbe R., Schaefer B., Wagner S., Mittermaier M., Machleidt F., Garcia C., Ruwwe-Glosenkamp C., Lingscheid T., Bosquillon de Jarcy L., Stegemann M. S., Pfeiffer M., Jurgens L., Denker S., Zickler D., Enghard P., Zelezniak A., Campbell A., Hayward C., Porteous D. J., Marioni R. E., Uhrig A., Muller-Redetzky H., Zoller H., Loffler-Ragg J., Keller M. A., Tancevski I., Timms J. F., Zaikin A., Hippenstiel S., Ramharter M., Witzenrath M., Suttorp N., Lilley K., Mulleder M., Sander L. E.; PA-COVID-19 Study group, Ralser M., Kurth F., A time-resolved proteomic and prognostic map of COVID-19. Cell Syst. 12, 780–794.e7 (2021). - PMC - PubMed
-
- Shu T., Ning W., Wu D., Xu J., Han Q., Huang M., Zou X., Yang Q., Yuan Y., Bie Y., Pan S., Mu J., Han Y., Yang X., Zhou H., Li R., Ren Y., Chen X., Yao S., Qiu Y., Zhang D. Y., Xue Y., Shang Y., Zhou X., Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. Immunity 53, 1108–1122.e5 (2020). - PMC - PubMed
MeSH terms
Substances
Grants and funding
- U19 AI090023/AI/NIAID NIH HHS/United States
- U19 AI128913/AI/NIAID NIH HHS/United States
- U19 AI118608/AI/NIAID NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U19 AI062629/AI/NIAID NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- R01 AI104870/AI/NIAID NIH HHS/United States
- U24 AI152179/AI/NIAID NIH HHS/United States
- R01 AG071858/AG/NIA NIH HHS/United States
- U19 AI125357/AI/NIAID NIH HHS/United States
- R01 AI145835/AI/NIAID NIH HHS/United States
- R01 AI135803/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

