Function investigation of p11.5 in ASFV infection
- PMID: 38789040
- PMCID: PMC11279770
- DOI: 10.1016/j.virs.2024.05.007
Function investigation of p11.5 in ASFV infection
Abstract
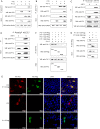
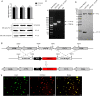
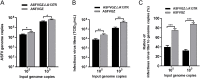
Virus replication relies on complex interactions between viral proteins. In the case of African swine fever virus (ASFV), only a few such interactions have been identified so far. In this study, we demonstrate that ASFV protein p72 interacts with p11.5 using co-immunoprecipitation and liquid chromatography-mass spectrometry (LC-MS). It was found that protein p72 interacts specifically with p11.5 at sites amino acids (aa) 1-216 of p72 and aa 1-68 of p11.5. To assess the importance of p11.5 in ASFV infection, we developed a recombinant virus (ASFVGZΔA137R) by deleting the A137R gene from the ASFVGZ genome. Compared with ASFVGZ, the infectious progeny virus titers of ASFVGZΔA137R were reduced by approximately 1.0 logs. In addition, we demonstrated that the growth defect was partially attributable to a higher genome copies-to-infectious virus titer ratios produced in ASFVGZΔA137R-infected MA104 cells than in those infected with ASFVGZ. This finding suggests that MA104 cells infected with ASFVGZΔA137R may generate larger quantities of noninfectious particles. Importantly, we found that p11.5 did not affect virus-cell binding or endocytosis. Collectively, we show for the first time the interaction between ASFV p72 and p11.5. Our results effectively provide the relevant information of the p11.5 protein. These results extend our understanding of complex interactions between viral proteins, paving the way for further studies of the potential mechanisms and pathogenesis of ASFV infection.
Keywords: African swine fever virus; Protein interactions; p11.5 protein; p72 protein.
Copyright © 2024 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest All authors declare that there are no competing interests.
Figures

Similar articles
-
Deletion of the H240R Gene of African Swine Fever Virus Decreases Infectious Progeny Virus Production Due to Aberrant Virion Morphogenesis and Enhances Inflammatory Cytokine Expression in Porcine Macrophages.J Virol. 2022 Feb 9;96(3):e0166721. doi: 10.1128/JVI.01667-21. Epub 2021 Nov 17. J Virol. 2022. PMID: 34787458 Free PMC article.
-
Insights into the Role of VPS39 and Its Interaction with CP204L and A137R in ASFV Infection.Viruses. 2024 Sep 17;16(9):1478. doi: 10.3390/v16091478. Viruses. 2024. PMID: 39339953 Free PMC article.
-
TRIM28 regulates the coagulation cascade inhibited by p72 of African swine fever virus.Vet Res. 2024 Nov 12;55(1):149. doi: 10.1186/s13567-024-01407-6. Vet Res. 2024. PMID: 39533356 Free PMC article.
-
Structure of African Swine Fever Virus and Associated Molecular Mechanisms Underlying Infection and Immunosuppression: A Review.Front Immunol. 2021 Sep 6;12:715582. doi: 10.3389/fimmu.2021.715582. eCollection 2021. Front Immunol. 2021. PMID: 34552586 Free PMC article. Review.
-
Cell entry mechanisms of African swine fever virus.Virology. 2024 Dec;600:110277. doi: 10.1016/j.virol.2024.110277. Epub 2024 Oct 24. Virology. 2024. PMID: 39488059 Review.
Cited by
-
Aloperine Inhibits ASFV via Regulating PRLR/JAK2 Signaling Pathway In Vitro.Int J Mol Sci. 2024 Aug 21;25(16):9083. doi: 10.3390/ijms25169083. Int J Mol Sci. 2024. PMID: 39201769 Free PMC article.
-
Punicalagin Inhibits African Swine Fever Virus Replication by Targeting Early Viral Stages and Modulating Inflammatory Pathways.Vet Sci. 2024 Sep 19;11(9):440. doi: 10.3390/vetsci11090440. Vet Sci. 2024. PMID: 39330819 Free PMC article.
References
-
- Alcami A., Angulo A., Vinuela E. Mapping and sequence of the gene encoding the African swine fever virion protein of M(r) 11500. J. Gen. Virol. 1993;74:2317–2324. - PubMed
-
- Borca M.V., Ramirez-Medina E., Silva E., Vuono E., Rai A., Pruitt S., Holinka L.G., Velazquez-Salinas L., Zhu J., Gladue D.P. Development of a highly effective African swine fever virus vaccine by deletion of the I177L gene results in sterile immunity against the current epidemic Eurasia strain. J. Virol. 2020;94 19. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

