A comparative analysis reveals the genomic diversity among 8 Muscovy duck populations
- PMID: 38789099
- PMCID: PMC11228869
- DOI: 10.1093/g3journal/jkae112
A comparative analysis reveals the genomic diversity among 8 Muscovy duck populations
Abstract
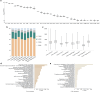
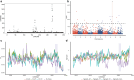
The Muscovy duck (Cairina moschata) is a waterfowl indigenous to the neotropical regions of Central and South America. It has low demand for concentrated feed and strong adaptability to different rearing conditions. After introduced to China through Eurasian commercial trade, Muscovy ducks have a domestication history of around 300 years in the Fujian Province of China. In the 1990s, the commodity Muscovy duck breed "Crimo," cultivated in Europe, entered the Chinese market for consumption and breeding purposes. Due to the different selective breeding processes, Muscovy ducks have various populational traits and lack transparency of their genetic background. To remove this burden in the Muscovy duck breeding process, we analyzed genomic data from 8 populations totaling 83 individuals. We identify 11.24 million single nucleotide polymorphisms (SNPs) and categorized these individuals into the Fujian-bred and the Crimo populations according to phylogenetic analyses. We then delved deeper into their evolutionary relationships through assessing population structure, calculating fixation index (FST) values, and measuring genetic distances. Our exploration of runs of homozygosity (ROHs) and homozygous-by-descent (HBD) uncovered genomic regions enriched for genes implicated in fatty acid metabolism, development, and immunity pathways. Selective sweep analyses further indicated strong selective pressures exerted on genes including TECR, STAT2, and TRAF5. These findings provide insights into genetic variations of Muscovy ducks, thus offering valuable information regarding genetic diversity, population conservation, and genome associated with the breeding of Muscovy ducks.
Keywords: SNPs; homozygous-by-descent; runs of homozygosity; selective sweep.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflicts of interest.
Figures



Similar articles
-
Analysis of genetic structure and identification of important genes associated with muscle growth in Fujian Muscovy duck.Poult Sci. 2024 Dec;103(12):104445. doi: 10.1016/j.psj.2024.104445. Epub 2024 Oct 28. Poult Sci. 2024. PMID: 39504826 Free PMC article.
-
Whole-genome selection signature differences between Chaohu and Ji'an red ducks.BMC Genomics. 2024 May 27;25(1):522. doi: 10.1186/s12864-024-10339-6. BMC Genomics. 2024. PMID: 38802792 Free PMC article.
-
Genomic variation responding to artificial selection on different lines of Pekin duck.Poult Sci. 2025 Feb;104(2):104785. doi: 10.1016/j.psj.2025.104785. Epub 2025 Jan 7. Poult Sci. 2025. PMID: 39813863 Free PMC article.
-
Genetic diversity and population structure of muscovy duck (Cairina moschata) from Nigeria.PeerJ. 2022 Apr 15;10:e13236. doi: 10.7717/peerj.13236. eCollection 2022. PeerJ. 2022. PMID: 35444865 Free PMC article.
-
Genetic diversity and relationship of Indian Muscovy duck populations.Mitochondrial DNA A DNA Mapp Seq Anal. 2018 Mar;29(2):165-169. doi: 10.1080/24701394.2016.1261851. Epub 2016 Dec 30. Mitochondrial DNA A DNA Mapp Seq Anal. 2018. PMID: 28034341
Cited by
-
Whole-genome resequencing reveals genetic differentiation and selection signatures among wild, local and commercial duck populations.Anim Biosci. 2025 May;38(5):898-909. doi: 10.5713/ab.24.0643. Epub 2024 Nov 28. Anim Biosci. 2025. PMID: 40336248 Free PMC article.
References
-
- Bencharit S, Zeng T, Jiang X, Li J, Wang D, Li G, Lu L, Wang G. 2013. Comparative proteomic analysis of the hepatic response to heat stress in Muscovy and Pekin ducks: insight into thermal tolerance related to energy metabolism. PLoS One. 8(10):e76917.. doi:10.1371/journal.pone.0076917. - DOI - PMC - PubMed
-
- Bertrand AR, Kadri NK, Flori L, Gautier M, Druet T, Leder E. 2019. RZooRoH: an R package to characterize individual genomic autozygosity and identify homozygous-by-descent segments. Methods Ecol Evol. 10(6):860–866. doi:10.1111/2041-210X.13167. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
