Validation of human telomere length multi-ancestry meta-analysis association signals identifies POP5 and KBTBD6 as human telomere length regulation genes
- PMID: 38789417
- PMCID: PMC11126610
- DOI: 10.1038/s41467-024-48394-y
Validation of human telomere length multi-ancestry meta-analysis association signals identifies POP5 and KBTBD6 as human telomere length regulation genes
Abstract
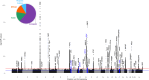
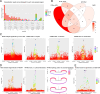
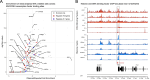
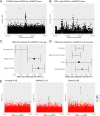
Genome-wide association studies (GWAS) have become well-powered to detect loci associated with telomere length. However, no prior work has validated genes nominated by GWAS to examine their role in telomere length regulation. We conducted a multi-ancestry meta-analysis of 211,369 individuals and identified five novel association signals. Enrichment analyses of chromatin state and cell-type heritability suggested that blood/immune cells are the most relevant cell type to examine telomere length association signals. We validated specific GWAS associations by overexpressing KBTBD6 or POP5 and demonstrated that both lengthened telomeres. CRISPR/Cas9 deletion of the predicted causal regions in K562 blood cells reduced expression of these genes, demonstrating that these loci are related to transcriptional regulation of KBTBD6 and POP5. Our results demonstrate the utility of telomere length GWAS in the identification of telomere length regulation mechanisms and validate KBTBD6 and POP5 as genes affecting telomere length regulation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: Juan C. Celedon received inhaled steroids from Merck for an NIH_funded study, unrelated to this work. Ivana V. Yang is a consultant for Eleven P15, a company focused on the early diagnosis and treatment of lung fibrosis. Dr. Patrick T. Ellinor receives sponsored research support from Bayer AG, IBM Research, Bristol Myers Squibb and Pfizer; he has also served on advisory boards or consulted for Bayer AG, MyoKardia and Novartis. Dr. David Schwartz is a founder and chief scientific officer of Eleven P15, a company focused on the early diagnosis and treatment of lung fibrosis. Laura M. Raffield is a consultant for the TOPMed Admistrative Coordinating Center (through Westat). Alexis Battle is a shareholder in Alphabet, Inc.; consultant for Third Rock Ventures, LLC; and founder of CellCipher, Inc. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
- P30 AG028747/AG/NIA NIH HHS/United States
- R01 DK107786/DK/NIDDK NIH HHS/United States
- U54 MD007597/MD/NIMHD NIH HHS/United States
- R01AG069120/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- U01 AG058589/AG/NIA NIH HHS/United States
- U01 HL072518/HL/NHLBI NIH HHS/United States
- R01 HL143818/HL/NHLBI NIH HHS/United States
- P01 HL162607/HL/NHLBI NIH HHS/United States
- UG1 HL139125/HL/NHLBI NIH HHS/United States
- R01 HG010297/HG/NHGRI NIH HHS/United States
- R01 HL079915/HL/NHLBI NIH HHS/United States
- R01 HL149836/HL/NHLBI NIH HHS/United States
- K12 GM123914/GM/NIGMS NIH HHS/United States
- R01 HL112064/HL/NHLBI NIH HHS/United States
- R01 ES021801/ES/NIEHS NIH HHS/United States
- R01HL153805/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01AG081244/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 HL139731/HL/NHLBI NIH HHS/United States
- P30 ES007048/ES/NIEHS NIH HHS/United States
- 5K12GM123914/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 AG081244/AG/NIA NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- R01 HL087698/HL/NHLBI NIH HHS/United States
- R01 ES016535/ES/NIEHS NIH HHS/United States
- R35CA209974/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R35 CA209974/CA/NCI NIH HHS/United States
- R01 HL076647/HL/NHLBI NIH HHS/United States
- R01HL105756/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 HL092577/HL/NHLBI NIH HHS/United States
- R01HL68959/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01HL079915/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 HL158668/HL/NHLBI NIH HHS/United States
- P01 ES009581/ES/NIEHS NIH HHS/United States
- R01 HL130974/HL/NHLBI NIH HHS/United States
- R01HL87681/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- P01 ES022845/ES/NIEHS NIH HHS/United States
- R01 HL061768/HL/NHLBI NIH HHS/United States
- R01 HL153805/HL/NHLBI NIH HHS/United States
- P50 CA180905/CA/NCI NIH HHS/United States
- R01HL-120393/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 AG058921/AG/NIA NIH HHS/United States
- U01 HL153009/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R35 HL135818/HL/NHLBI NIH HHS/United States
- I01 BX005295/BX/BLRD VA/United States
- RC2 HL101651/HL/NHLBI NIH HHS/United States
- P01 ES011627/ES/NIEHS NIH HHS/United States
- R01 AG069120/AG/NIA NIH HHS/United States
- R35GM139580/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- UM1 AI160040/AI/NIAID NIH HHS/United States
- R01 HL087680/HL/NHLBI NIH HHS/United States
- M01 RR000052/RR/NCRR NIH HHS/United States
- R00 ES027870/ES/NIEHS NIH HHS/United States
- R01 AI132476/AI/NIAID NIH HHS/United States
- U01 HL137162/HL/NHLBI NIH HHS/United States
- R01 AI153239/AI/NIAID NIH HHS/United States
- R01 GM152471/GM/NIGMS NIH HHS/United States
- U01 AG052409/AG/NIA NIH HHS/United States
- R01 ES023262/ES/NIEHS NIH HHS/United States
- R01 HL068959/HL/NHLBI NIH HHS/United States
- R03 ES014046/ES/NIEHS NIH HHS/United States
- R01 DK071891/DK/NIDDK NIH HHS/United States
- R35 GM139580/GM/NIGMS NIH HHS/United States
- U01 AI160018/AI/NIAID NIH HHS/United States
- R01 HL157635/HL/NHLBI NIH HHS/United States
- R01 HL087681/HL/NHLBI NIH HHS/United States
- UG3 HL151865/HL/NHLBI NIH HHS/United States

