Constitutional mismatch repair deficiency mimicking Lynch syndrome is associated with hypomorphic mismatch repair gene variants
- PMID: 38789506
- PMCID: PMC11126593
- DOI: 10.1038/s41698-024-00603-z
Constitutional mismatch repair deficiency mimicking Lynch syndrome is associated with hypomorphic mismatch repair gene variants
Abstract
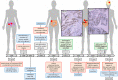
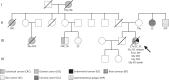
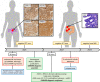
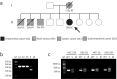
Lynch syndrome (LS) and constitutional mismatch repair deficiency (CMMRD) are distinct cancer syndromes caused, respectively, by mono- and bi-allelic germline mismatch repair (MMR) variants. LS predisposes to mainly gastrointestinal and genitourinary cancers in adulthood. CMMRD predisposes to brain, haematological, and LS-spectrum cancers from childhood. Two suspected LS patients with first cancer diagnosis aged 27 or 38 years were found to be homozygous for an MMR (likely) pathogenic variant, MSH6 c.3226C>T (p.(Arg1076Cys)), or variant of uncertain significance (VUS), MLH1 c.306G>A (p.(Glu102=)). MLH1 c.306G>A was shown to cause leaky exon 3 skipping. The apparent genotype-phenotype conflict was resolved by detection of constitutional microsatellite instability in both patients, a hallmark feature of CMMRD. A hypomorphic effect of these and other variants found in additional late onset CMMRD cases, identified by literature review, likely explains a LS-like phenotype. CMMRD testing in carriers of compound heterozygous or homozygous MMR VUS may find similar cases and novel hypomorphic variants. Individualised management of mono- and bi-allelic carriers of hypomorphic MMR variants is needed until we better characterise the associated phenotypes.
© 2024. The Author(s).
Conflict of interest statement
RG and JB are named co-inventors on a patent covering the microsatellite instability markers analysed in the cMSI assay: GB2114136.1 (filed October 1, 2021). All other authors declare no financial or non-financial competing interests.
Figures
Similar articles
-
Germline mismatch repair gene mutations in children with tumors: a case series from two centers.Transl Pediatr. 2024 Oct 1;13(10):1810-1819. doi: 10.21037/tp-24-343. Epub 2024 Oct 28. Transl Pediatr. 2024. PMID: 39524392 Free PMC article.
-
Germline variants screening of MLH1, MSH2, MSH6 and PMS2 genes in 64 Algerian Lynch syndrome families: The first nationwide study.Ann Hum Genet. 2022 Nov;86(6):328-352. doi: 10.1111/ahg.12482. Epub 2022 Sep 8. Ann Hum Genet. 2022. PMID: 36073783
-
Constitutional Microsatellite Instability, Genotype, and Phenotype Correlations in Constitutional Mismatch Repair Deficiency.Gastroenterology. 2023 Apr;164(4):579-592.e8. doi: 10.1053/j.gastro.2022.12.017. Epub 2022 Dec 29. Gastroenterology. 2023. PMID: 36586540
-
[Constitutional MMR deficiency: Genetic bases and clinical implications].Bull Cancer. 2019 Feb;106(2):162-172. doi: 10.1016/j.bulcan.2018.10.008. Epub 2018 Dec 11. Bull Cancer. 2019. PMID: 30551794 Review. French.
-
Diagnostic criteria for constitutional mismatch repair deficiency syndrome: suggestions of the European consortium 'care for CMMRD' (C4CMMRD).J Med Genet. 2014 Jun;51(6):355-65. doi: 10.1136/jmedgenet-2014-102284. Epub 2014 Apr 15. J Med Genet. 2014. PMID: 24737826 Review.
Cited by
-
Genetics, genomics and clinical features of adenomatous polyposis.Fam Cancer. 2025 Apr 16;24(2):38. doi: 10.1007/s10689-025-00460-0. Fam Cancer. 2025. PMID: 40237887 Free PMC article. Review.
-
The CRISPR-Cas System and Clinical Applications of CRISPR-Based Gene Editing in Hematology with a Focus on Inherited Germline Predisposition to Hematologic Malignancies.Genes (Basel). 2024 Jul 1;15(7):863. doi: 10.3390/genes15070863. Genes (Basel). 2024. PMID: 39062641 Free PMC article. Review.
-
Unraveling mutagenic processes influencing the tumor mutational patterns of individuals with constitutional mismatch repair deficiency.Nat Commun. 2025 May 14;16(1):4459. doi: 10.1038/s41467-025-59775-2. Nat Commun. 2025. PMID: 40368937 Free PMC article.
-
ERN GENTURIS guidelines on constitutional mismatch repair deficiency diagnosis, genetic counselling, surveillance, quality of life, and clinical management.Eur J Hum Genet. 2024 Dec;32(12):1526-1541. doi: 10.1038/s41431-024-01708-6. Epub 2024 Oct 17. Eur J Hum Genet. 2024. PMID: 39420201 Free PMC article. Review.
-
Lynch syndrome and colorectal cancer: A review of current perspectives in molecular genetics and clinical strategies.Oncol Res. 2025 Jun 26;33(7):1531-1545. doi: 10.32604/or.2025.063951. eCollection 2025. Oncol Res. 2025. PMID: 40612862 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

