Tead4 and Tfap2c generate bipotency and a bistable switch in totipotent embryos to promote robust lineage diversification
- PMID: 38789684
- PMCID: PMC11189297
- DOI: 10.1038/s41594-024-01311-9
Tead4 and Tfap2c generate bipotency and a bistable switch in totipotent embryos to promote robust lineage diversification
Abstract
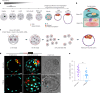
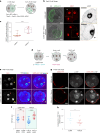
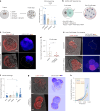
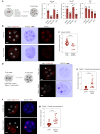
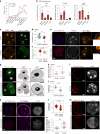
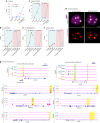
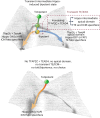
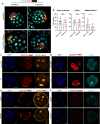
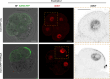
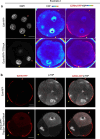
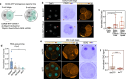
The mouse and human embryo gradually loses totipotency before diversifying into the inner cell mass (ICM, future organism) and trophectoderm (TE, future placenta). The transcription factors TFAP2C and TEAD4 with activated RHOA accelerate embryo polarization. Here we show that these factors also accelerate the loss of totipotency. TFAP2C and TEAD4 paradoxically promote and inhibit Hippo signaling before lineage diversification: they drive expression of multiple Hippo regulators while also promoting apical domain formation, which inactivates Hippo. Each factor activates TE specifiers in bipotent cells, while TFAP2C also activates specifiers of the ICM fate. Asymmetric segregation of the apical domain reconciles the opposing regulation of Hippo signaling into Hippo OFF and the TE fate, or Hippo ON and the ICM fate. We propose that the bistable switch established by TFAP2C and TEAD4 is exploited to trigger robust lineage diversification in the developing embryo.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

