A molecular glue RBM39-degrader induces synthetic lethality in cancer cells with homologous recombination repair deficiency
- PMID: 38789724
- PMCID: PMC11126574
- DOI: 10.1038/s41698-024-00610-0
A molecular glue RBM39-degrader induces synthetic lethality in cancer cells with homologous recombination repair deficiency
Abstract
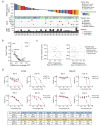
E7820 and Indisulam (E7070) are sulfonamide molecular glues that modulate RNA splicing by degrading the splicing factor RBM39 via ternary complex formation with the E3 ligase adaptor DCAF15. To identify biomarkers of the antitumor efficacy of E7820, we treated patient-derived xenograft (PDX) mouse models established from 42 patients with solid tumors. The overall response rate was 38.1% (16 PDXs), and tumor regression was observed across various tumor types. Exome sequencing of the PDX genome revealed that loss-of-function mutations in genes of the homologous recombination repair (HRR) system, such as ATM, were significantly enriched in tumors that responded to E7820 (p = 4.5 × 103). Interestingly, E7820-mediated double-strand breaks in DNA were increased in tumors with BRCA2 dysfunction, and knockdown of BRCA1/2 transcripts or knockout of ATM, ATR, or BAP1 sensitized cancer cells to E7820. Transcriptomic analyses revealed that E7820 treatment resulted in the intron retention of mRNAs and decreased transcription, especially for HRR genes. This induced HRR malfunction probably leads to the synthetic lethality of tumor cells with homologous recombination deficiency (HRD). Furthermore, E7820, in combination with olaparib, exerted a synergistic effect, and E7820 was even effective in an olaparib-resistant cell line. In conclusion, HRD is a promising predictive biomarker of E7820 efficacy and has a high potential to improve the prognosis of patients with HRD-positive cancers.
© 2024. The Author(s).
Conflict of interest statement
N.Y. reports research grants from Astellas, Chugai, Eisai, Taiho, BMS, Pfizer, Novartis, Eli Lilly, AbbVie, Daiichi-Sankyo, Bayer, Boehringer Ingelheim, Kyowa-Hakko Kirin, Takeda, ONO, Janssen Pharma, MSD, MERCK, GSK, Sumitomo Dainippon, Chiome Bioscience, Otsuka, Carna Biosciences, Genmab, Shionogi, TORAY, KAKEN, InventisBio, and Rakuten Medical outside the submitted work as well as advisory fee from Eisai, Takeda, Boehringer Ingelheim, Cimic, and Chugai, and honoraria from ONO, Chugai, Daiichi-Sankyo, and Eisai outside the submitted work. K. Toshimitsu, K. Tabata, A.Y., and T.O. are Eisai employees.
Figures



References
-
- Funahashi Y, et al. Sulfonamide derivative, E7820, is a unique angiogenesis inhibitor suppressing an expression of integrin alpha2 subunit on endothelium. Cancer Res. 2002;62:6116–6123. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

