Objectivizing issues in the diagnosis of complex rare diseases: lessons learned from testing existing diagnosis support systems on ciliopathies
- PMID: 38789985
- PMCID: PMC11127295
- DOI: 10.1186/s12911-024-02538-8
Objectivizing issues in the diagnosis of complex rare diseases: lessons learned from testing existing diagnosis support systems on ciliopathies
Abstract
Background: There are approximately 8,000 different rare diseases that affect roughly 400 million people worldwide. Many of them suffer from delayed diagnosis. Ciliopathies are rare monogenic disorders characterized by a significant phenotypic and genetic heterogeneity that raises an important challenge for clinical diagnosis. Diagnosis support systems (DSS) applied to electronic health record (EHR) data may help identify undiagnosed patients, which is of paramount importance to improve patients' care. Our objective was to evaluate three online-accessible rare disease DSSs using phenotypes derived from EHRs for the diagnosis of ciliopathies.
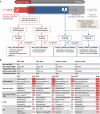
Methods: Two datasets of ciliopathy cases, either proven or suspected, and two datasets of controls were used to evaluate the DSSs. Patient phenotypes were automatically extracted from their EHRs and converted to Human Phenotype Ontology terms. We tested the ability of the DSSs to diagnose cases in contrast to controls based on Orphanet ontology.
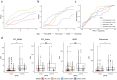
Results: A total of 79 cases and 38 controls were selected. Performances of the DSSs on ciliopathy real world data (best DSS with area under the ROC curve = 0.72) were not as good as published performances on the test set used in the DSS development phase. None of these systems obtained results which could be described as "expert-level". Patients with multisystemic symptoms were generally easier to diagnose than patients with isolated symptoms. Diseases easily confused with ciliopathy generally affected multiple organs and had overlapping phenotypes. Four challenges need to be considered to improve the performances: to make the DSSs interoperable with EHR systems, to validate the performances in real-life settings, to deal with data quality, and to leverage methods and resources for rare and complex diseases.
Conclusion: Our study provides insights into the complexities of diagnosing highly heterogenous rare diseases and offers lessons derived from evaluation existing DSSs in real-world settings. These insights are not only beneficial for ciliopathy diagnosis but also hold relevance for the enhancement of DSS for various complex rare disorders, by guiding the development of more clinically relevant rare disease DSSs, that could support early diagnosis and finally make more patients eligible for treatment.
Keywords: Artificial intelligence; Ciliopathy; Clinical decision support; Early diagnosis; Electronic health record; External evaluation; Human phenotype ontology; Patient similarity; Rare diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- RARE Disease Facts. Global Genes. https://globalgenes.org/rare-disease-facts/. Cited 2022 Jul 8.
-
- Escudié JB, Rance B, Malamut G, Khater S, Burgun A, Cellier C, et al. A novel data-driven workflow combining literature and electronic health records to estimate comorbidities burden for a specific disease: a case study on autoimmune comorbidities in patients with celiac disease. BMC Med Inf Decis Mak. 2017;17:140. doi: 10.1186/s12911-017-0537-y. - DOI - PMC - PubMed
MeSH terms
Grants and funding
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- ANR-17-RHUS-0002/Agence Nationale de la Recherche
- PE 3135/1-1/Deutsche Forschungsgemeinschaft
LinkOut - more resources
Full Text Sources
Medical

