Prenatal Genome-Wide Cell-Free DNA Screening: Three Years of Clinical Experience in a Hospital Prenatal Diagnostic Unit in Spain
- PMID: 38790198
- PMCID: PMC11121632
- DOI: 10.3390/genes15050568
Prenatal Genome-Wide Cell-Free DNA Screening: Three Years of Clinical Experience in a Hospital Prenatal Diagnostic Unit in Spain
Abstract
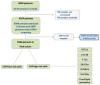
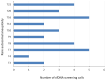
Genome-wide prenatal cell-free DNA (cfDNA) screening can be used to screen for a wide range of fetal chromosomal anomalies in pregnant patients. In this study, we describe our clinical experience with a genome-wide cfDNA assay in screening for common trisomies, sex chromosomal aneuploidies (SCAs), rare autosomal aneuploidies (RAAs), and copy-number variations (CNVs) in about 6000 patients over a three-year period at our hospital's Prenatal Diagnostic Unit in Spain. Overall, 204 (3.3%) patients had a high-risk call, which included 76 trisomy 21, 21 trisomy 18, 7 trisomy 13, 29 SCAs, 31 RAAs, 31 CNVs, and 9 cases with multiple anomalies. The diagnostic outcomes were obtained for the high-risk cases when available, allowing for the calculation of positive predictive values (PPVs). Calculated PPVs were 95.9% for trisomy 21, 77.8% for trisomy 18, 66.7% for trisomy 13, 10.7% for RAAs, and 10.7% for CNVs. Pregnancy and birth outcomes were also collected for the majority of RAA and CNV cases. Adverse perinatal outcomes for some of these cases included preeclampsia, fetal growth restriction, preterm birth, reduced birth weight, and major congenital structural abnormalities. In conclusion, our study showed strong performance for genome-wide cfDNA screening in a large cohort of pregnancy patients in Spain.
Keywords: copy-number variations; genome-wide; prenatal cfDNA screening; rare autosomal aneuploidies; trisomy.
Conflict of interest statement
Pedrola reports receiving open access publication fee and in-kind editorial support from Instituto de Investigación Sanitaria La Fe (IISLAFE) through a grant from Illumina, Inc. The authors declare no other conflicts of interest.
Figures
References
-
- Audibert F., De Bie I., Johnson J.A., Okun N., Wilson R.D., Armour C., Chitayat D., Kim R. No. 348-Joint SOGC-CCMG Guideline: Update on Prenatal Screening for Fetal Aneuploidy, Fetal Anomalies, and Adverse Pregnancy Outcomes. J. Obstet. Gynaecol. Can. 2017;39:805–817. doi: 10.1016/j.jogc.2017.01.032. - DOI - PubMed
-
- Dondorp W., de Wert G., Bombard Y., Bianchi D.W., Bergmann C., Borry P., Chitty L.S., Fellmann F., Forzano F., Hall A., et al. Non-invasive prenatal testing for aneuploidy and beyond: Challenges of responsible innovation in prenatal screening. Eur. J. Hum. Genet. 2015;23:1438–1450. doi: 10.1038/ejhg.2015.57. - DOI - PMC - PubMed
-
- Dungan J.S., Klugman S., Darilek S., Malinowski J., Akkari Y.M.N., Monaghan K.G., Erwin A., Best R.G. Noninvasive prenatal screening (NIPS) for fetal chromosome abnormalities in a general-risk population: An evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2023;25:100336. doi: 10.1016/j.gim.2022.11.004. - DOI - PubMed
-
- Gil Mira M.d.M., Santacruz Martín B., Molina García F.S., Paco Matallana K.d. Decalogue of the lc-DNA test in maternal blood for prenatal diagnosis (2020) Prog. Obstet. Ginecol. (Ed. Impr.) 2020;63:1–2. doi: 10.20960/j.pog.002. - DOI
-
- Hui L., Ellis K., Mayen D., Pertile M.D., Reimers R., Sun L., Vermeesch J., Vora N.L., Chitty L.S. Position statement from the International Society for Prenatal Diagnosis on the use of non-invasive prenatal testing for the detection of fetal chromosomal conditions in singleton pregnancies. Prenat. Diagn. 2023;43:814–828. doi: 10.1002/pd.6357. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

