Rare Genetic Developmental Disabilities: Mabry Syndrome (MIM 239300) Index Cases and Glycophosphatidylinositol (GPI) Disorders
- PMID: 38790248
- PMCID: PMC11121671
- DOI: 10.3390/genes15050619
Rare Genetic Developmental Disabilities: Mabry Syndrome (MIM 239300) Index Cases and Glycophosphatidylinositol (GPI) Disorders
Abstract
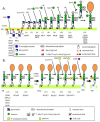
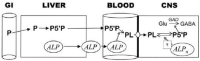
The case report by Mabry et al. (1970) of a family with four children with elevated tissue non-specific alkaline phosphatase, seizures and profound developmental disability, became the basis for phenotyping children with the features that became known as Mabry syndrome. Aside from improvements in the services available to patients and families, however, the diagnosis and treatment of this, and many other developmental disabilities, did not change significantly until the advent of massively parallel sequencing. As more patients with features of the Mabry syndrome were identified, exome and genome sequencing were used to identify the glycophosphatidylinositol (GPI) biosynthesis disorders (GPIBDs) as a group of congenital disorders of glycosylation (CDG). Biallelic variants of the phosphatidylinositol glycan (PIG) biosynthesis, type V (PIGV) gene identified in Mabry syndrome became evidence of the first in a phenotypic series that is numbered HPMRS1-6 in the order of discovery. HPMRS1 [MIM: 239300] is the phenotype resulting from inheritance of biallelic PIGV variants. Similarly, HPMRS2 (MIM 614749), HPMRS5 (MIM 616025) and HPMRS6 (MIM 616809) result from disruption of the PIGO, PIGW and PIGY genes expressed in the endoplasmic reticulum. By contrast, HPMRS3 (MIM 614207) and HPMRS4 (MIM 615716) result from disruption of post attachment to proteins PGAP2 (HPMRS3) and PGAP3 (HPMRS4). The GPI biosynthesis disorders (GPIBDs) are currently numbered GPIBD1-21. Working with Dr. Mabry, in 2020, we were able to use improved laboratory diagnostics to complete the molecular diagnosis of patients he had originally described in 1970. We identified biallelic variants of the PGAP2 gene in the first reported HPMRS patients. We discuss the longevity of the Mabry syndrome index patients in the context of the utility of pyridoxine treatment of seizures and evidence for putative glycolipid storage in patients with HPMRS3. From the perspective of the laboratory innovations made that enabled the identification of the HPMRS phenotype in Dr. Mabry's patients, the need for treatment innovations that will benefit patients and families affected by developmental disabilities is clear.
Keywords: Mabry syndrome; case study; glycophosphatidylinositol (GPI) biosynthesis disorder (GPIBD); human phenotype ontology (HPO) analysis; hyperphosphatasia with neurologic deficit (HPMRS); identity by descent filtering; whole exon sequencing; whole genome sequencing.
Conflict of interest statement
The author declares no conflicts of interest.
Figures




References
-
- Arteche-López A., Gómez Rodríguez M.J., Sánchez Calvin M.T., Quesada-Espinosa J.F., Lezana Rosales J.M., Palma Milla C., Gómez-Manjón I., Hidalgo Mayoral I., Pérez de la Fuente R., Díaz de Bustamante A., et al. Towards a Change in the Diagnostic Algorithm of Autism Spectrum Disorders: Evidence Supporting Whole Exome Sequencing as a First-Tier Test. Genes. 2021;12:560. doi: 10.3390/genes12040560. - DOI - PMC - PubMed
-
- Sun Y., Peng J., Liang D., Ye X., Xu N., Chen L., Yan D., Zhang H., Xiao B., Qiu W., et al. Genome sequencing demonstrates high diagnostic yield in children with undiagnosed global developmental delay/intellectual disability: A prospective study. Hum. Mutat. 2022;43:568–581. doi: 10.1002/humu.24347. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

