Puerarin Modulates Hepatic Farnesoid X Receptor and Gut Microbiota in High-Fat Diet-Induced Obese Mice
- PMID: 38791314
- PMCID: PMC11121391
- DOI: 10.3390/ijms25105274
Puerarin Modulates Hepatic Farnesoid X Receptor and Gut Microbiota in High-Fat Diet-Induced Obese Mice
Abstract
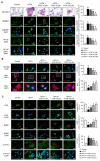
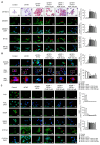
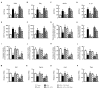
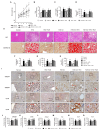
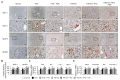
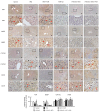
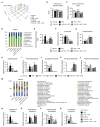
Obesity is associated with alterations in lipid metabolism and gut microbiota dysbiosis. This study investigated the effects of puerarin, a bioactive isoflavone, on lipid metabolism disorders and gut microbiota in high-fat diet (HFD)-induced obese mice. Supplementation with puerarin reduced plasma alanine aminotransferase, liver triglyceride, liver free fatty acid (FFA), and improved gut microbiota dysbiosis in obese mice. Puerarin's beneficial metabolic effects were attenuated when farnesoid X receptor (FXR) was antagonized, suggesting FXR-mediated mechanisms. In hepatocytes, puerarin ameliorated high FFA-induced sterol regulatory element-binding protein (SREBP) 1 signaling, inflammation, and mitochondrial dysfunction in an FXR-dependent manner. In obese mice, puerarin reduced liver damage, regulated hepatic lipogenesis, decreased inflammation, improved mitochondrial function, and modulated mitophagy and ubiquitin-proteasome pathways, but was less effective in FXR knockout mice. Puerarin upregulated hepatic expression of FXR, bile salt export pump (BSEP), and downregulated cytochrome P450 7A1 (CYP7A1) and sodium taurocholate transporter (NTCP), indicating modulation of bile acid synthesis and transport. Puerarin also restored gut microbial diversity, the Firmicutes/Bacteroidetes ratio, and the abundance of Clostridium celatum and Akkermansia muciniphila. This study demonstrates that puerarin effectively ameliorates metabolic disturbances and gut microbiota dysbiosis in obese mice, predominantly through FXR-dependent pathways. These findings underscore puerarin's potential as a therapeutic agent for managing obesity and enhancing gut health, highlighting its dual role in improving metabolic functions and modulating microbial communities.
Keywords: farnesoid X receptor; gut microbiota; mitochondrial function; mitophagy; obese; puerarin.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Farnesoid X Receptor: Effective alleviation of rifampicin -induced liver injury.Int Immunopharmacol. 2024 Sep 30;139:112799. doi: 10.1016/j.intimp.2024.112799. Epub 2024 Jul 27. Int Immunopharmacol. 2024. PMID: 39068755
-
FXR Signaling-Mediated Bile Acid Metabolism Is Critical for Alleviation of Cholesterol Gallstones by Lactobacillus Strains.Microbiol Spectr. 2022 Oct 26;10(5):e0051822. doi: 10.1128/spectrum.00518-22. Epub 2022 Aug 29. Microbiol Spectr. 2022. PMID: 36036629 Free PMC article.
-
Tauroursodeoxycholic Acid Improves Nonalcoholic Fatty Liver Disease by Regulating Gut Microbiota and Bile Acid Metabolism.J Agric Food Chem. 2024 Sep 11;72(36):20194-20210. doi: 10.1021/acs.jafc.4c04630. Epub 2024 Aug 28. J Agric Food Chem. 2024. PMID: 39193771
-
An Intestinal Microbiota-Farnesoid X Receptor Axis Modulates Metabolic Disease.Gastroenterology. 2016 Nov;151(5):845-859. doi: 10.1053/j.gastro.2016.08.057. Epub 2016 Sep 14. Gastroenterology. 2016. PMID: 27639801 Free PMC article. Review.
-
Bile acid-microbiota crosstalk in hepatitis B virus infection.J Gastroenterol Hepatol. 2024 Aug;39(8):1509-1516. doi: 10.1111/jgh.16604. Epub 2024 May 9. J Gastroenterol Hepatol. 2024. PMID: 38721685 Review.
Cited by
-
Puerarin as a multi-targeted modulator of lipid metabolism: molecular mechanisms, therapeutic potential and prospects for nutritional translation.Front Nutr. 2025 Jul 18;12:1598897. doi: 10.3389/fnut.2025.1598897. eCollection 2025. Front Nutr. 2025. PMID: 40756564 Free PMC article. Review.
-
Advances in the acting mechanism and treatment of gut microbiota in metabolic dysfunction-associated steatotic liver disease.Gut Microbes. 2025 Dec;17(1):2500099. doi: 10.1080/19490976.2025.2500099. Epub 2025 May 20. Gut Microbes. 2025. PMID: 40394806 Free PMC article. Review.
-
Liver Cell Mitophagy in Metabolic Dysfunction-Associated Steatotic Liver Disease and Liver Fibrosis.Antioxidants (Basel). 2024 Jun 15;13(6):729. doi: 10.3390/antiox13060729. Antioxidants (Basel). 2024. PMID: 38929168 Free PMC article. Review.
-
Unveiling the power of Pueraria lobata: a comprehensive exploration of its medicinal and edible potentials.Front Pharmacol. 2025 Jul 10;16:1578472. doi: 10.3389/fphar.2025.1578472. eCollection 2025. Front Pharmacol. 2025. PMID: 40709088 Free PMC article. Review.
-
Gut microbiota-mitochondrial crosstalk in obesity: novel mechanistic insights and therapeutic strategies with traditional Chinese medicine.Front Pharmacol. 2025 Apr 22;16:1574887. doi: 10.3389/fphar.2025.1574887. eCollection 2025. Front Pharmacol. 2025. PMID: 40331200 Free PMC article. Review.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

