Clinical and Immunologic Characteristics of Colorectal Cancer Tumors Expressing LY6G6D
- PMID: 38791382
- PMCID: PMC11121234
- DOI: 10.3390/ijms25105345
Clinical and Immunologic Characteristics of Colorectal Cancer Tumors Expressing LY6G6D
Abstract
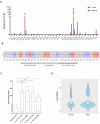
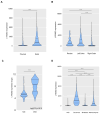
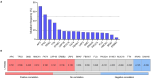
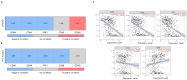
The identification of targets that are expressed on the cell membrane is a main goal in cancer research. The Lymphocyte Antigen 6 Family Member G6D (LY6G6D) gene codes for a protein that is mainly present on the surface of colorectal cancer (CRC) cells. Therapeutic strategies against this protein like the development of T cell engagers (TCE) are currently in the early clinical stage. In the present work, we interrogated public genomic datasets including TCGA to evaluate the genomic and immunologic cell profile present in tumors with high expression of LY6G6D. We used data from TCGA, among others, and the Tumor Immune Estimation Resource (TIMER2.0) platform for immune cell estimations and Spearman correlation tests. LY6G6D expression was exclusively present in CRC, particularly in the microsatellite stable (MSS) subtype, and was associated with left-side tumors and the canonical genomic subgroup. Tumors with mutations of APC and p53 expressed elevated levels of LY6G6D. This protein was expressed in tumors with an inert immune microenvironment with an absence of immune cells and co-inhibitory molecules. In conclusion, we described clinical, genomic and immune-pathologic characteristics that can be used to optimize the clinical development of agents against this target. Future studies should be performed to confirm these findings and potentially explore the suggested clinical development options.
Keywords: CRC; LY6G6D; TCEs; immune association.
Conflict of interest statement
Alberto Ocaña declares current consulting fees from NMS, and previously from real-world clinical trials, viewpoint, Daichii, Entrechem, CancerAppy, Servier and Symphogen. Former employee of Symphogen. Founder and president of the not-for-profit cancer research foundation ACEPAIN. Victor Moreno declares consulting fees from Roche, Bayer, BMS, Janssen, Syneos, Affimed and AstraZeneca. Principal Investigator—Institutional Funding: AbbVie, AceaBio, Adaptimmune, ADC Therapeutics, Aduro, Agenus, Amcure, Amgen, Astellas, AstraZeneca Bayer Beigene BioInvent International AB, BMS, Boehringer, Boheringer, Boston, Celgene, Daichii Sankyo, DEBIOPHARM, Eisai, e-Terapeutics, Exelisis, Forma Therapeutics, Genmab, GSK, Harpoon, Hutchison, Immutep, Incyte, Inovio, Iovance, Janssen, Kyowa Kirin, Lilly, Loxo, MedSir, Menarini, Merck, Merus, Millennium, MSD, Nanobiotix, Nektar, Novartis, Odonate Therapeutics, Pfizer, PharmaMar, Principia, PsiOxus, Puma, Regeneron, Rigontec, Roche, Sanofi, Sierra Oncology, Synthon, Taiho, Takeda, Tesaro, Transgene, Turning Point Therapeutics, Upshersmith. Emiliano Calvo declares advisory board participation (financial interest) for Adcendo, Alkermes, Amunix, Anaveon, Amcure, AstraZeneca, BMS, Janssen, MonTa, MSD, Nanobiotix, Nouscom, Novartis, OncoDNA, PharmaMar, Roche/Genentech, Sanofi, Servier, SyneosHealth, TargImmune, and T-knife; research grants for Achilles and BeiGene; steering committee (financial interest) and IDMC participation for BeiGene (IDMC steering committee), EORTC IDMC chair (non-financial interest), MedSIR (steering committee), and Novartis (steering committee); honoraria for scientific board participation for Adcendo, Chugai Pharmaceuticals, and PsiOxus Therapeutics; employment at HM Hospitals Group and START Program of Early Phase Clinical Drug Development in Oncology, START corporation, Oncoart Associated, and International Cancer Consultants and is the founder and president of the not-for-profit Investigational Therapeutics in Oncological Sciences (INTHEOS) Foundation. There are no conflicts of interest to declare in relation to this manuscript. The rest of the authors declare that they have no competing interests.
Figures


Similar articles
-
Decoding LY6G6D in colorectal cancer: Unraveling biomarker potential and therapeutic insights.Cell Mol Biol (Noisy-le-grand). 2024 Jun 5;70(6):14-20. doi: 10.14715/cmb/2024.70.6.3. Cell Mol Biol (Noisy-le-grand). 2024. PMID: 38836687
-
JAK/Stat5-mediated subtype-specific lymphocyte antigen 6 complex, locus G6D (LY6G6D) expression drives mismatch repair proficient colorectal cancer.J Exp Clin Cancer Res. 2019 Jan 22;38(1):28. doi: 10.1186/s13046-018-1019-5. J Exp Clin Cancer Res. 2019. PMID: 30670049 Free PMC article.
-
LY6G6D is a selectively expressed colorectal cancer antigen that can be used for targeting a therapeutic T-cell response by a T-cell engager.Front Immunol. 2022 Sep 8;13:1008764. doi: 10.3389/fimmu.2022.1008764. eCollection 2022. Front Immunol. 2022. PMID: 36159851 Free PMC article.
-
Potential Therapeutic Targets of B7 Family in Colorectal Cancer.Front Immunol. 2020 May 5;11:681. doi: 10.3389/fimmu.2020.00681. eCollection 2020. Front Immunol. 2020. PMID: 32477326 Free PMC article. Review.
-
Role of adenomatous polyposis coli (APC) gene mutations in the pathogenesis of colorectal cancer; current status and perspectives.Biochimie. 2019 Feb;157:64-71. doi: 10.1016/j.biochi.2018.11.003. Epub 2018 Nov 8. Biochimie. 2019. PMID: 30414835 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

