Substrate Affinity Is Not Crucial for Therapeutic L-Asparaginases: Antileukemic Activity of Novel Bacterial Enzymes
- PMID: 38792133
- PMCID: PMC11124013
- DOI: 10.3390/molecules29102272
Substrate Affinity Is Not Crucial for Therapeutic L-Asparaginases: Antileukemic Activity of Novel Bacterial Enzymes
Abstract
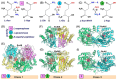
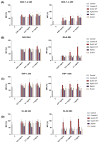
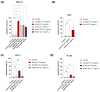
L-asparaginases are used in the treatment of acute lymphoblastic leukemia. The aim of this work was to compare the antiproliferative potential and proapoptotic properties of novel L-asparaginases from different structural classes, viz. EcAIII and KpAIII (class 2), as well as ReAIV and ReAV (class 3). The EcAII (class 1) enzyme served as a reference. The proapoptotic and antiproliferative effects were tested using four human leukemia cell models: MOLT-4, RAJI, THP-1, and HL-60. The antiproliferative assay with the MOLT-4 cell line indicated the inhibitory properties of all tested L-asparaginases. The results from the THP-1 cell models showed a similar antiproliferative effect in the presence of EcAII, EcAIII, and KpAIII. In the case of HL-60 cells, the inhibition of proliferation was observed in the presence of EcAII and KpAIII, whereas the proliferation of RAJI cells was inhibited only by EcAII. The results of the proapoptotic assays showed individual effects of the enzymes toward specific cell lines, suggesting a selective (time-dependent and dose-dependent) action of the tested L-asparaginases. We have, thus, demonstrated that novel L-asparaginases, with a lower substrate affinity than EcAII, also exhibit significant antileukemic properties in vitro, which makes them interesting new drug candidates for the treatment of hematological malignancies. For all enzymes, the kinetic parameters (Km and kcat) and thermal stability (Tm) were determined. Structural and catalytic properties of L-asparaginases from different classes are also summarized.
Keywords: L-asparaginase; cell apoptosis; cell proliferation; leukemia; substrate affinity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
L-Asparaginase from Lachancea Thermotolerans: Effect of Lys99Ala on Enzyme Performance and in vitro Antileukemic Efficacy.Biotechnol J. 2024 Nov;19(11):e202400507. doi: 10.1002/biot.202400507. Biotechnol J. 2024. PMID: 39552048
-
Structural and biochemical properties of L-asparaginase.FEBS J. 2021 Jul;288(14):4183-4209. doi: 10.1111/febs.16042. Epub 2021 Jun 19. FEBS J. 2021. PMID: 34060231 Review.
-
[Bacterial recombinant L-asparaginases: properties, structure and anti-proliferative activity].Biomed Khim. 2015 May-Jun;61(3):312-24. doi: 10.18097/PBMC20156103312. Biomed Khim. 2015. PMID: 26215408 Review. Russian.
-
Rhizobium etli has two L-asparaginases with low sequence identity but similar structure and catalytic center.Acta Crystallogr D Struct Biol. 2023 Aug 1;79(Pt 8):775-791. doi: 10.1107/S2059798323005648. Epub 2023 Jul 26. Acta Crystallogr D Struct Biol. 2023. PMID: 37494066
-
The E. coli L-asparaginase V27T mutant: structural and functional characterization and comparison with theoretical predictions.FEBS Lett. 2022 Dec;596(23):3060-3068. doi: 10.1002/1873-3468.14526. Epub 2022 Nov 7. FEBS Lett. 2022. PMID: 36310372 Free PMC article.
References
-
- Broome J.D. Evidence That the L-Asparaginase Activity of Guinea Pig Serum Is Responsible for Its Antilymphoma Effects. Nature. 1961;191:1114–1115. doi: 10.1038/1911114a0. - DOI
-
- Karamitros C.S., Labrou N.E. Extracellular Expression and Affinity Purification of L-Asparaginase from E. chrysanthemi in E. coli. Sustain. Chem. Process. 2014;2:16. doi: 10.1186/s40508-014-0016-z. - DOI
-
- Van Trimpont M., Peeters E., De Visser Y., Schalk A.M., Mondelaers V., De Moerloose B., Lavie A., Lammens T., Goossens S., Van Vlierberghe P. Novel Insights on the Use of L-Asparaginase as an Efficient and Safe Anti-Cancer Therapy. Cancers. 2022;14:902. doi: 10.3390/cancers14040902. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

