Genomic Characterization and Molecular Detection of Rehmannia Allexivirus Virus, a Novel Allexivirus Infecting Rehmannia glutinosa
- PMID: 38792674
- PMCID: PMC11123084
- DOI: 10.3390/microorganisms12050844
Genomic Characterization and Molecular Detection of Rehmannia Allexivirus Virus, a Novel Allexivirus Infecting Rehmannia glutinosa
Abstract
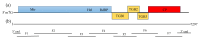
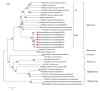
Rehmannia glutinosa is one of the most important medicinal plants in China and is affected by viral diseases. In this study, a new virus tentatively named Rehmannia Allexivirus virus (ReAV) was identified through high-throughput sequencing, reverse-transcription polymerase chain reaction (RT-PCR), and Sanger sequencing. The complete genome length was 7297 nt and it contained five open reading frames (ORFs) encoding replicase, triple gene block 1(TGB1), TGB2, TGB3, and coat protein (CP). The replicase and CP presented nucleotide homology ranges of 59.9-65.2% and 47.5-55.5% between the nine ReAV isolates and the other 12 species of the genus Allexivirus. In the nine isolates, ReAV-20 and ReAV-31 isolates showed breakpoints in the replicase and CP regions, respectively. The other isolates shared 87.2-96.5% nt with the whole genome nucleotide identity. The phylogenetic tree showed that seven ReAV isolates based on replicase, CP, and whole genome sequences were clustered in the same branch and were related to the genus Allexivirus. The ReAV detection rates for 60 R. glutinosa samples were 73.3-81.7% through RT-PCR using primers targeting the replicase or CP genes. These results demonstrate that ReAV is the dominant virus in R. glutinosa. This study provides important evidence for understanding viruses infecting R. glutinosa and for establishing efficient strategies to prevent viral spread.
Keywords: Allexivirus; ReAV; genomic sequence; molecular variation; phylogenetic relationship.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Zhang B., Li X., Wang F., Li M., Zhang J., Gu L., Zhang L., Tu W., Zhang Z. Assaying the potential autotoxins and microbial community associated with Rehmannia glutinosa replant problems based on its ‘autotoxic circle’. Plant Soil. 2016;407:307–322. doi: 10.1007/s11104-016-2885-2. - DOI
-
- Du J.F., Nian W.K., Zhou Z.J., Dou T., Song G.H., Gu L., Liu X.Y. Leaf Spot Disease Caused by Alternaria alternata on Rehmannia glutinosa in China. Plant Dis. 2020;104:3059. doi: 10.1094/PDIS-01-20-0108-PDN. - DOI
-
- Ahn S.W., Kim Y.G., Kim M.H., Lee H.Y., Seong N.S. Comparison of biological activities on Rehmannia radix and R. radix Preparata produced in Korea. Korean J. Med. Crop Sci. 1999;7:257–262.
Grants and funding
- CARS-21/China Agriculture Research System of MOF and MARA
- HARS-22-11-Z1/Henan Modern Industrial Technology System of Chinese Herbal Medicine
- 2023XK05/the New Discipline Development Project of Henan Academy of Agricultural Sciences
- 2023ZC051/independent innovation project of Henan Academy of Agricultural Sciences
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

