From Investigating a Case of Cellulitis to Exploring Nosocomial Infection Control of ST1 Legionella pneumophila Using Genomic Approaches
- PMID: 38792686
- PMCID: PMC11123157
- DOI: 10.3390/microorganisms12050857
From Investigating a Case of Cellulitis to Exploring Nosocomial Infection Control of ST1 Legionella pneumophila Using Genomic Approaches
Abstract
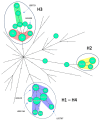
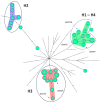
Legionella pneumophila can cause a large panel of symptoms besides the classic pneumonia presentation. Here we present a case of fatal nosocomial cellulitis in an immunocompromised patient followed, a year later, by a second case of Legionnaires' disease in the same ward. While the first case was easily assumed as nosocomial based on the date of symptom onset, the second case required clear typing results to be assigned either as nosocomial and related to the same environmental source as the first case, or community acquired. To untangle this specific question, we applied core-genome multilocus typing (MLST), whole-genome single nucleotide polymorphism and whole-genome MLST methods to a collection of 36 Belgian and 41 international sequence-type 1 (ST1) isolates using both thresholds recommended in the literature and tailored threshold based on local epidemiological data. Based on the thresholds applied to cluster isolates together, the three methods gave different results and no firm conclusion about the nosocomial setting of the second case could been drawn. Our data highlight that despite promising results in the study of outbreaks and for large-scale epidemiological investigations, next-generation sequencing typing methods applied to ST1 outbreak investigation still need standardization regarding both wet-lab protocols and bioinformatics. A deeper evaluation of the L. pneumophila evolutionary clock is also required to increase our understanding of genomic differences between isolates sampled during a clinical infection and in the environment.
Keywords: Legionella pneumophila; Legionella pneumophila ST1; cgMLST; genomic typing; hygiene investigation; nosocomial; wgMLST; wgSNP; whole genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Wüthrich D., Gautsch S., Spieler-Denz R., Dubuis O., Gaia V., Moran-Gilad J., Hinic V., Seth-Smith H.M., Nickel C.H., Tschudin-Sutter S., et al. Air-conditioner cooling towers as complex reservoirs and continuous source of Legionella pneumophila infection evidenced by a genomic analysis study in 2017, Switzerland. Eurosurveillance. 2019;24:1800192. doi: 10.2807/1560-7917.ES.2019.24.4.1800192. - DOI - PMC - PubMed
-
- Ginevra C., Chastang J., David S., Mentasti M., Yakunin E., Chalker V.J., Chalifa-Caspi V., Valinsky L., Jarraud S., Moran-Gilad J. A real-time PCR for specific detection of the Legionella pneumophila serogroup 1 ST1 complex. Clin. Microbiol. Infect. 2020;26:514.e1–514.e6. doi: 10.1016/j.cmi.2019.09.002. - DOI - PubMed
-
- Echahidi F.M.C. Activity Report from 2011 to 2022 Reference Centre for Legionella pneumophila UZ Brussel—LHUB-ULB [Internet]. National Reference Centre for Legionella pneumophila, Vrije Universiteit Brussel (VUB), Universitair Ziekenhuis Brussel (UZ Brussel), Department of Microbiology and Infection Control, Brussels. [(accessed on 16 April 2024)]. Available online: https://www.sciensano.be/sites/default/files/legionella_2011-2022_nrc_ra....
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

