The Impact of Aboveground Epichloë Endophytic Fungi on the Rhizosphere Microbial Functions of the Host Melica transsilvanica
- PMID: 38792786
- PMCID: PMC11124418
- DOI: 10.3390/microorganisms12050956
The Impact of Aboveground Epichloë Endophytic Fungi on the Rhizosphere Microbial Functions of the Host Melica transsilvanica
Abstract
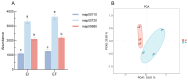
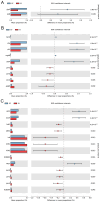
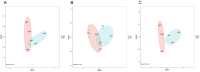
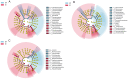
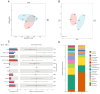
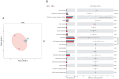
In nature, the symbiotic relationship between plants and microorganisms is crucial for ecosystem balance and plant growth. This study investigates the impact of Epichloë endophytic fungi, which are exclusively present aboveground, on the rhizosphere microbial functions of the host Melica transsilvanica. Using metagenomic methods, we analyzed the differences in microbial functional groups and functional genes in the rhizosphere soil between symbiotic (EI) and non-symbiotic (EF) plants. The results reveal that the presence of Epichloë altered the community structure of carbon and nitrogen cycling-related microbial populations in the host's rhizosphere, significantly increasing the abundance of the genes (porA, porG, IDH1) involved in the rTCA cycle of the carbon fixation pathway, as well as the abundance of nxrAB genes related to nitrification in the nitrogen-cycling pathway. Furthermore, the presence of Epichloë reduces the enrichment of virulence factors in the host rhizosphere microbiome, while significantly increasing the accumulation of resistance genes against heavy metals such as Zn, Sb, and Pb. This study provides new insights into the interactions among endophytic fungi, host plants, and rhizosphere microorganisms, and offers potential applications for utilizing endophytic fungi resources to improve plant growth and soil health.
Keywords: Epichloë; Melica transsilvanica; metagenomics; rhizosphere microbial functions; symbiosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Compant S., Clément C., Sessitsch A. Plant growth-promoting bacteria in the rhizo-and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010;42:669–678. doi: 10.1016/j.soilbio.2009.11.024. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

