Design and Development of an Antigen Test for SARS-CoV-2 Nucleocapsid Protein to Validate the Viral Quality Assurance Panels
- PMID: 38793544
- PMCID: PMC11125937
- DOI: 10.3390/v16050662
Design and Development of an Antigen Test for SARS-CoV-2 Nucleocapsid Protein to Validate the Viral Quality Assurance Panels
Abstract
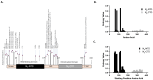
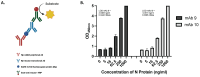
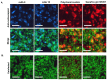
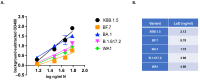
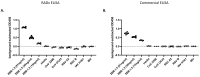
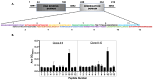
The continuing mutability of the SARS-CoV-2 virus can result in failures of diagnostic assays. To address this, we describe a generalizable bioinformatics-to-biology pipeline developed for the calibration and quality assurance of inactivated SARS-CoV-2 variant panels provided to Radical Acceleration of Diagnostics programs (RADx)-radical program awardees. A heuristic genetic analysis based on variant-defining mutations demonstrated the lowest genetic variance in the Nucleocapsid protein (Np)-C-terminal domain (CTD) across all SARS-CoV-2 variants. We then employed the Shannon entropy method on (Np) sequences collected from the major variants, verifying the CTD with lower entropy (less prone to mutations) than other Np regions. Polyclonal and monoclonal antibodies were raised against this target CTD antigen and used to develop an Enzyme-linked immunoassay (ELISA) test for SARS-CoV-2. Blinded Viral Quality Assurance (VQA) panels comprised of UV-inactivated SARS-CoV-2 variants (XBB.1.5, BF.7, BA.1, B.1.617.2, and WA1) and distractor respiratory viruses (CoV 229E, CoV OC43, RSV A2, RSV B, IAV H1N1, and IBV) were assembled by the RADx-rad Diagnostics core and tested using the ELISA described here. The assay tested positive for all variants with high sensitivity (limit of detection: 1.72-8.78 ng/mL) and negative for the distractor virus panel. Epitope mapping for the monoclonal antibodies identified a 20 amino acid antigenic peptide on the Np-CTD that an in-silico program also predicted for the highest antigenicity. This work provides a template for a bioinformatics pipeline to select genetic regions with a low propensity for mutation (low Shannon entropy) to develop robust 'pan-variant' antigen-based assays for viruses prone to high mutational rates.
Keywords: COVID-19 diagnostics; Enzyme-linked immunoassay; RADx; SARS-CoV-2; monoclonal and polyclonal antibodies; nucleocapsid protein; peptide epitope mapping; viral quality assurance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- RADxSM Radical. [(accessed on 8 March 2024)]. Available online: https://www.radxrad.org/
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

