Towards Understanding and Identification of Human Viral Co-Infections
- PMID: 38793555
- PMCID: PMC11126107
- DOI: 10.3390/v16050673
Towards Understanding and Identification of Human Viral Co-Infections
Abstract
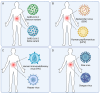
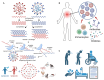
Viral co-infections, in which a host is infected with multiple viruses simultaneously, are common in the human population. Human viral co-infections can lead to complex interactions between the viruses and the host immune system, affecting the clinical outcome and posing challenges for treatment. Understanding the types, mechanisms, impacts, and identification methods of human viral co-infections is crucial for the prevention and control of viral diseases. In this review, we first introduce the significance of studying human viral co-infections and summarize the current research progress and gaps in this field. We then classify human viral co-infections into four types based on the pathogenic properties and species of the viruses involved. Next, we discuss the molecular mechanisms of viral co-infections, focusing on virus-virus interactions, host immune responses, and clinical manifestations. We also summarize the experimental and computational methods for the identification of viral co-infections, emphasizing the latest advances in high-throughput sequencing and bioinformatics approaches. Finally, we highlight the challenges and future directions in human viral co-infection research, aiming to provide new insights and strategies for the prevention, control, diagnosis, and treatment of viral diseases. This review provides a comprehensive overview of the current knowledge and future perspectives on human viral co-infections and underscores the need for interdisciplinary collaboration to address this complex and important topic.
Keywords: clinical symptoms; experimental and computational identification; host immune response; human viral co-infections; viral interactions.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Ryan F.P. Viruses as symbionts. Symbiosis. 2007;44:11–21.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

