Development of a Cell Culture Model for Inducible SARS-CoV-2 Replication
- PMID: 38793589
- PMCID: PMC11125939
- DOI: 10.3390/v16050708
Development of a Cell Culture Model for Inducible SARS-CoV-2 Replication
Abstract
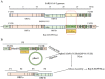
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces direct cytopathic effects, complicating the establishment of low-cytotoxicity cell culture models for studying its replication. We initially developed a DNA vector-based replicon system utilizing the CMV promoter to generate a recombinant viral genome bearing reporter genes. However, this system frequently resulted in drug resistance and cytotoxicity, impeding model establishment. Herein, we present a novel cell culture model with SARS-CoV-2 replication induced by Cre/LoxP-mediated DNA recombination. An engineered SARS-CoV-2 transcription unit was subcloned into a bacterial artificial chromosome (BAC) vector. To enhance biosafety, the viral spike protein gene was deleted, and the nucleocapsid gene was replaced with a reporter gene. An exogenous sequence was inserted within NSP1 as a modulatory cassette that is removable after Cre/LoxP-mediated DNA recombination and subsequent RNA splicing. Using the PiggyBac transposon strategy, the transcription unit was integrated into host cell chromatin, yielding a stable cell line capable of inducing recombinant SARS-CoV-2 RNA replication. The model exhibited sensitivity to the potential antivirals forsythoside A and verteporfin. An innovative inducible SARS-CoV-2 replicon cell model was introduced to further explore the replication and pathogenesis of the virus and facilitate screening and assessment of anti-SARS-CoV-2 therapeutics.
Keywords: inducible model; replicon; severe acute respiratory syndrome coronavirus 2.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

