Epigenetic Modifications of White Blood Cell DNA Caused by Transient Fetal Infection with Bovine Viral Diarrhea Virus
- PMID: 38793603
- PMCID: PMC11125956
- DOI: 10.3390/v16050721
Epigenetic Modifications of White Blood Cell DNA Caused by Transient Fetal Infection with Bovine Viral Diarrhea Virus
Abstract
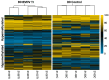
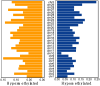
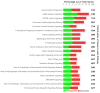
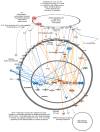
Bovine viral diarrhea virus (BVDV) infections cause USD 1.5-2 billion in losses annually. Maternal BVDV after 150 days of gestation causes transient fetal infection (TI) in which the fetal immune response clears the virus. The impact of fetal TI BVDV infections on postnatal growth and white blood cell (WBC) methylome as an index of epigenetic modifications was examined by inoculating pregnant heifers with noncytopathic type 2 BVDV or media (sham-inoculated controls) on Day 175 of gestation to generate TI (n = 11) and control heifer calves (n = 12). Fetal infection in TI calves was confirmed by virus-neutralizing antibody titers at birth and control calves were seronegative. Both control and TI calves were negative for BVDV RNA in WBCs by RT-PCR. The mean weight of the TI calves was less than that of the controls (p < 0.05). DNA methyl seq analysis of WBC DNA demonstrated 2349 differentially methylated cytosines (p ≤ 0.05) including 1277 hypomethylated cytosines, 1072 hypermethylated cytosines, 84 differentially methylated regions based on CpGs in promoters, and 89 DMRs in islands of TI WBC DNA compared to controls. Fetal BVDV infection during late gestation resulted in epigenomic modifications predicted to affect fetal development and immune pathways, suggesting potential consequences for postnatal growth and health of TI cattle.
Keywords: bovine viral diarrhea virus; epigenetics; immune system; intrauterine growth restriction; pathway analysis; transient infection.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

