Long-Read Nanopore-Based Sequencing of Anelloviruses
- PMID: 38793605
- PMCID: PMC11125752
- DOI: 10.3390/v16050723
Long-Read Nanopore-Based Sequencing of Anelloviruses
Abstract
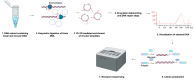
Routinely used metagenomic next-generation sequencing (mNGS) techniques often fail to detect low-level viremia (<104 copies/mL) and appear biased towards viruses with linear genomes. These limitations hinder the capacity to comprehensively characterize viral infections, such as those attributed to the Anelloviridae family. These near ubiquitous non-pathogenic components of the human virome have circular single-stranded DNA genomes that vary in size from 2.0 to 3.9 kb and exhibit high genetic diversity. Hence, species identification using short reads can be challenging. Here, we introduce a rolling circle amplification (RCA)-based metagenomic sequencing protocol tailored for circular single-stranded DNA genomes, utilizing the long-read Oxford Nanopore platform. The approach was assessed by sequencing anelloviruses in plasma drawn from people who inject drugs (PWID) in two geographically distinct cohorts. We detail the methodological adjustments implemented to overcome difficulties inherent in sequencing circular genomes and describe a computational pipeline focused on anellovirus detection. We assessed our protocol across various sample dilutions and successfully differentiated anellovirus sequences in conditions simulating mixed infections. This method provides a robust framework for the comprehensive characterization of circular viruses within the human virome using the Oxford Nanopore.
Keywords: bioinformatics; circular viral genomes; metagenomics; plasma; rolling circle amplification; virome.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Arze C.A., Springer S., Dudas G., Patel S., Bhattacharyya A., Swaminathan H., Brugnara C., Delagrave S., Ong T., Kahvejian A., et al. Global genome analysis reveals a vast and dynamic anellovirus landscape within the human virome. Cell Host Microbe. 2021;29:1305–1315.e1306. doi: 10.1016/j.chom.2021.07.001. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

