User-Friendly Replication-Competent MAdV-1 Vector System with a Cloning Capacity of 3.3 Kilobases
- PMID: 38793642
- PMCID: PMC11126015
- DOI: 10.3390/v16050761
User-Friendly Replication-Competent MAdV-1 Vector System with a Cloning Capacity of 3.3 Kilobases
Abstract
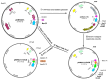
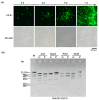
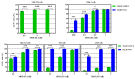
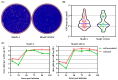
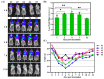
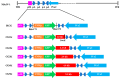
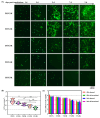
Mouse adenoviruses (MAdV) play important roles in studying host-adenovirus interaction. However, easy-to-use reverse genetics systems are still lacking for MAdV. An infectious plasmid pKRMAV1 was constructed by ligating genomic DNA of wild-type MAdV-1 with a PCR product containing a plasmid backbone through Gibson assembly. A fragment was excised from pKRMAV1 by restriction digestion and used to generate intermediate plasmid pKMAV1-ER, which contained E3, fiber, E4, and E1 regions of MAdV-1. CMV promoter-controlled GFP expression cassette was inserted downstream of the pIX gene in pKMAV1-ER and then transferred to pKRMAV1 to generate adenoviral plasmid pKMAV1-IXCG. Replacement of transgene could be conveniently carried out between dual BstZ17I sites in pKMAV1-IXCG by restriction-assembly, and a series of adenoviral plasmids were generated. Recombinant viruses were rescued after transfecting linearized adenoviral plasmids to mouse NIH/3T3 cells. MAdV-1 viruses carrying GFP or firefly luciferase genes were characterized in gene transduction, plaque-forming, and replication in vitro or in vivo by observing the expression of reporter genes. The results indicated that replication-competent vectors presented relevant properties of wild-type MAdV-1 very well. By constructing viruses bearing exogenous fragments with increasing size, it was found that MAdV-1 could tolerate an insertion up to 3.3 kb. Collectively, a replication-competent MAdV-1 vector system was established, which simplified procedures for the change of transgene or modification of E1, fiber, E3, or E4 genes.
Keywords: Gibson assembly; infectious plasmid; mouse adenovirus 1; packaging capacity; replication competent; transgene; vector.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Site-directed modification of adenoviral vector with combined DNA assembly and restriction-ligation cloning.J Biotechnol. 2020 Jan 10;307:193-201. doi: 10.1016/j.jbiotec.2019.11.009. Epub 2019 Nov 18. J Biotechnol. 2020. PMID: 31751597
-
Single Plasmid-Based, Upgradable, and Backward-Compatible Adenoviral Vector Systems.Hum Gene Ther. 2019 Jun;30(6):777-791. doi: 10.1089/hum.2018.258. Epub 2019 Mar 27. Hum Gene Ther. 2019. PMID: 30793964
-
Restriction-Assembly: A Solution to Construct Novel Adenovirus Vector.Viruses. 2022 Mar 6;14(3):546. doi: 10.3390/v14030546. Viruses. 2022. PMID: 35336953 Free PMC article.
-
Construction of Adenoviral Vectors using DNA Assembly Technology.J Vis Exp. 2022 Jun 16;(184). doi: 10.3791/64033. J Vis Exp. 2022. PMID: 35786609
-
Efficient generation of double heterologous promoter controlled oncolytic adenovirus vectors by a single homologous recombination step in Escherichia coli.BMC Biotechnol. 2006 Aug 3;6:36. doi: 10.1186/1472-6750-6-36. BMC Biotechnol. 2006. PMID: 16887042 Free PMC article.
Cited by
-
Role of mouse adenovirus type 1 E4orf6-induced degradation of protein kinase R in pathogenesis.J Virol. 2025 Feb 25;99(2):e0154524. doi: 10.1128/jvi.01545-24. Epub 2024 Dec 31. J Virol. 2025. PMID: 39745442 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources

