Antigenic Characterization of Circulating and Emerging SARS-CoV-2 Variants in the U.S. throughout the Delta to Omicron Waves
- PMID: 38793756
- PMCID: PMC11125585
- DOI: 10.3390/vaccines12050505
Antigenic Characterization of Circulating and Emerging SARS-CoV-2 Variants in the U.S. throughout the Delta to Omicron Waves
Abstract
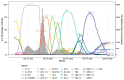
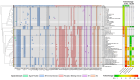
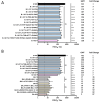
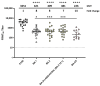
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has evolved into numerous lineages with unique spike mutations and caused multiple epidemics domestically and globally. Although COVID-19 vaccines are available, new variants with the capacity for immune evasion continue to emerge. To understand and characterize the evolution of circulating SARS-CoV-2 variants in the U.S., the Centers for Disease Control and Prevention (CDC) initiated the National SARS-CoV-2 Strain Surveillance (NS3) program and has received thousands of SARS-CoV-2 clinical specimens from across the nation as part of a genotype to phenotype characterization process. Focus reduction neutralization with various antisera was used to antigenically characterize 143 SARS-CoV-2 Delta, Mu and Omicron subvariants from selected clinical specimens received between May 2021 and February 2023, representing a total of 59 unique spike protein sequences. BA.4/5 subvariants BU.1, BQ.1.1, CR.1.1, CQ.2 and BA.4/5 + D420N + K444T; BA.2.75 subvariants BM.4.1.1, BA.2.75.2, CV.1; and recombinant Omicron variants XBF, XBB.1, XBB.1.5 showed the greatest escape from neutralizing antibodies when analyzed against post third-dose original monovalent vaccinee sera. Post fourth-dose bivalent vaccinee sera provided better protection against those subvariants, but substantial reductions in neutralization titers were still observed, especially among BA.4/5 subvariants with both an N-terminal domain (NTD) deletion and receptor binding domain (RBD) substitutions K444M + N460K and recombinant Omicron variants. This analysis demonstrated a framework for long-term systematic genotype to antigenic characterization of circulating and emerging SARS-CoV-2 variants in the U.S., which is critical to assessing their potential impact on the effectiveness of current vaccines and antigen recommendations for future updates.
Keywords: COVID-19 vaccine; Delta variant; Omicron variant; SARS-CoV-2; antigenic characterization; neutralizing antibody.
Conflict of interest statement
H.H was supported by Synergy America, Inc. and S.H.P. was supported by Eagle Global Scientific, Inc. Synergy America, Inc. and Eagle Global Scientific, Inc. had no involvement in study design; collection, analysis, and interpretation of data; writing of the report and the decision to submit the report for publication.
Figures


References
-
- Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks J.T., MacNeil A., Slayton R.B., Tong S., Silk B.J., Armstrong G.L., et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage–United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. - DOI - PMC - PubMed
-
- Paul P., France A.M., Aoki Y., Batra D., Biggerstaff M., Dugan V., Galloway S., Hall A.J., Johansson M.A., Kondor R.J., et al. Genomic Surveillance for SARS-CoV-2 Variants Circulating in the United States, December 2020–May 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:846–850. doi: 10.15585/mmwr.mm7023a3. - DOI - PMC - PubMed
-
- Lambrou A.S., Shirk P., Steele M.K., Paul P., Paden C.R., Cadwell B., Reese H.E., Aoki Y., Hassell N., Zheng X.Y., et al. Genomic Surveillance for SARS-CoV-2 Variants: Predominance of the Delta (B.1.617.2) and Omicron (B.1.1.529) Variants—United States, June 2021–January 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:206–211. doi: 10.15585/mmwr.mm7106a4. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

