A Hybrid Approach Combining Shape-Based and Docking Methods to Identify Novel Potential P2X7 Antagonists from Natural Product Databases
- PMID: 38794162
- PMCID: PMC11123696
- DOI: 10.3390/ph17050592
A Hybrid Approach Combining Shape-Based and Docking Methods to Identify Novel Potential P2X7 Antagonists from Natural Product Databases
Abstract
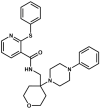
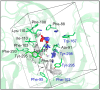
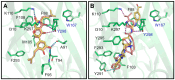
P2X7 is an ATP-activated purinergic receptor implicated in pro-inflammatory responses. It is associated with the development of several diseases, including inflammatory and neurodegenerative conditions. Although several P2X7 receptor antagonists have recently been reported in the literature, none of them is approved for clinical use. However, the structure of the known antagonists can serve as a scaffold for discovering effective compounds in clinical therapy. This study aimed to propose an improved virtual screening methodology for the identification of novel potential P2X7 receptor antagonists from natural products through the combination of shape-based and docking approaches. First, a shape-based screening was performed based on the structure of JNJ-47965567, a P2X7 antagonist, using two natural product compound databases, MEGx (~5.8 × 103 compounds) and NATx (~32 × 103 compounds). Then, the compounds selected by the proposed shape-based model, with Shape-Tanimoto score values ranging between 0.624 and 0.799, were filtered for drug-like properties. Finally, the compounds that met the drug-like filter criteria were docked into the P2X7 allosteric binding site, using the docking programs GOLD and DockThor. The docking poses with the best score values were submitted to careful visual inspection of the P2X7 allosteric binding site. Based on our established visual inspection criteria, four compounds from the MEGx database and four from the NATx database were finally selected as potential P2X7 receptor antagonists. The selected compounds are structurally different from known P2X7 antagonists, have drug-like properties, and are predicted to interact with key P2X7 allosteric binding pocket residues, including F88, F92, F95, F103, M105, F108, Y295, Y298, and I310. Therefore, the combination of shape-based screening and docking approaches proposed in our study has proven useful in selecting potential novel P2X7 antagonist candidates from natural-product-derived compounds databases. This approach could also be useful for selecting potential inhibitors/antagonists of other receptors and/or biological targets.
Keywords: P2X7 receptor; antagonists; natural products; shape-based model; virtual screening.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



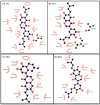
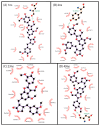

Similar articles
-
Screening herbal and natural product libraries to aid discovery of novel allosteric modulators of human P2X7.Purinergic Signal. 2025 Apr;21(2):365-379. doi: 10.1007/s11302-024-10055-6. Epub 2024 Oct 22. Purinergic Signal. 2025. PMID: 39436616 Free PMC article.
-
Structure-based virtual screening of the nociceptin receptor: hybrid docking and shape-based approaches for improved hit identification.J Chem Inf Model. 2014 Oct 27;54(10):2732-43. doi: 10.1021/ci500291a. Epub 2014 Sep 17. J Chem Inf Model. 2014. PMID: 25148595 Free PMC article.
-
Identification of novel P2X7R antagonists by using structure-based virtual screening and cell-based assays.Chem Biol Drug Des. 2021 Jul;98(1):192-205. doi: 10.1111/cbdd.13867. Epub 2021 Jun 3. Chem Biol Drug Des. 2021. PMID: 33993620
-
Structure-Activity Relationships and Therapeutic Potential of Purinergic P2X7 Receptor Antagonists.Curr Med Chem. 2024;31(11):1361-1403. doi: 10.2174/0929867330666230403094538. Curr Med Chem. 2024. PMID: 37013427 Review.
-
The purinergic P2X7 receptor as a potential drug target to combat neuroinflammation in neurodegenerative diseases.Med Res Rev. 2020 Nov;40(6):2427-2465. doi: 10.1002/med.21710. Epub 2020 Jul 16. Med Res Rev. 2020. PMID: 32677086 Review.
Cited by
-
Screening herbal and natural product libraries to aid discovery of novel allosteric modulators of human P2X7.Purinergic Signal. 2025 Apr;21(2):365-379. doi: 10.1007/s11302-024-10055-6. Epub 2024 Oct 22. Purinergic Signal. 2025. PMID: 39436616 Free PMC article.
-
Identification of New Human P2X7 Antagonists Using Ligand- and Structure-Based Virtual Screening.J Chem Inf Model. 2025 Jul 14;65(13):7143-7155. doi: 10.1021/acs.jcim.5c00552. Epub 2025 Jun 26. J Chem Inf Model. 2025. PMID: 40566963 Free PMC article.
References
-
- Alves L.A., de Melo Reis R.A., de Souza C.A.M., de Freitas M.S., Teixeira P.C.N., Neto Moreira Ferreira D., Xavier R.F. The P2X7 Receptor: Shifting from a Low- to a High-Conductance Channel—An Enigmatic Phenomenon? Biochim. Biophys. Acta (BBA)—Biomembr. 2014;1838:2578–2587. doi: 10.1016/j.bbamem.2014.05.015. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

