Small Molecule Drugs Targeting Viral Polymerases
- PMID: 38794231
- PMCID: PMC11124969
- DOI: 10.3390/ph17050661
Small Molecule Drugs Targeting Viral Polymerases
Abstract
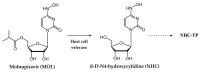
Small molecules that specifically target viral polymerases-crucial enzymes governing viral genome transcription and replication-play a pivotal role in combating viral infections. Presently, approved polymerase inhibitors cover nine human viruses, spanning both DNA and RNA viruses. This review provides a comprehensive analysis of these licensed drugs, encompassing nucleoside/nucleotide inhibitors (NIs), non-nucleoside inhibitors (NNIs), and mutagenic agents. For each compound, we describe the specific targeted virus and related polymerase enzyme, the mechanism of action, and the relevant bioactivity data. This wealth of information serves as a valuable resource for researchers actively engaged in antiviral drug discovery efforts, offering a complete overview of established strategies as well as insights for shaping the development of next-generation antiviral therapeutics.
Keywords: FDA; drugs; inhibitors; small molecules; viral polymerase; viruses.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




















References
-
- Choi K.H. Viral Molecular Machines. Springer; Boston, MA, USA: 2012. Viral polymerases; pp. 267–304. Book Series: Advances in Experimental Medicine and Biology (AEMB, volume 726) - DOI
Publication types
LinkOut - more resources
Full Text Sources

