Bt-Modified Transgenic Rice May Shift the Composition and Diversity of Rhizosphere Microbiota
- PMID: 38794371
- PMCID: PMC11125220
- DOI: 10.3390/plants13101300
Bt-Modified Transgenic Rice May Shift the Composition and Diversity of Rhizosphere Microbiota
Abstract
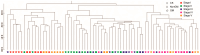
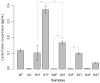
Plants significantly shape root-associated microbiota, making rhizosphere microbes useful environmental indicator organisms for safety assessment. Here, we report the pyrosequencing of the bacterial 16S ribosomal RNA in rhizosphere soil samples collected from transgenic cry1Ab/cry1Ac Bt rice Huahui No. 1 (GM crop) and its parental counterpart, Minghui63. We identified a total of 2579 quantifiable bacterial operational taxonomic units (OTUs). Many treatment-enriched microbial OTUs were identified, including 14 NonGM-enriched OTUs and 10 GM-enriched OTUs. OTUs belonging to the phyla Proteobacteria, Actinobacteria, Acidobacteria, Firmicutes, Nitrospirae, Chlorobi and GN04 were identified as statistically different in abundance between GM and the other two treatments. Compared with the different impacts of different rice varieties on microbiota, the impact of rice planting on microbiota is more obvious. Furthermore, Huahui No. 1 transgenic Bt rice had a greater impact on the rhizosphere bacterial communities than Minghui63. Early developmental stages of the transgenic Bt rice had a significant impact on many Bacillaceae communities. Soil chemical properties were not significantly altered by the presence of transgenic Bt rice. The peak concentration level of Bt protein products was detected during the seedling stage of transgenic Bt rice, which may be an intriguing factor for bacterial diversity variations. Based on these findings, we conclude that transgenic Bt rice has a significant impact on root-associated bacteria. This information may be leveraged in future environmental safety assessments of transgenic Bt rice varieties.
Keywords: 16S ribosomal RNA; genetically modified rice; microbial communities; operational taxonomic units; transgenic Bt rice variety.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Mishra P.K., Mishra S., Selvakumar G., Kundu S., Shankar Gupta H. Enhanced soybean (Glycine max L.) plant growth and nodulation by Bradyrhizobium japonicum-SB1 in presence of Bacillus thuringiensis-KR1. Acta Agric. Scand. Sect. B-Soil Plant Sci. 2009;59:189–196.
LinkOut - more resources
Full Text Sources

