Protocol for the development of mRNA lipid nanoparticle vaccines and analysis of immunization efficiency in mice
- PMID: 38795353
- PMCID: PMC11144802
- DOI: 10.1016/j.xpro.2024.103087
Protocol for the development of mRNA lipid nanoparticle vaccines and analysis of immunization efficiency in mice
Abstract
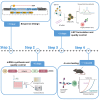
Here, we present a protocol for the development of mRNA-loaded lipid nanoparticle (LNP) vaccines for target antigen sequences of interest. We describe key steps required to design and synthesize mRNA constructs, their LNP encapsulation, and mouse immunization. We then detail quality control assays to determine RNA purity, guidelines to measure RNA immunogenicity using in vitro reporter systems, and a technique to evaluate antigen-specific T cell responses following immunization.
Keywords: biotechnology and bioengineering; cancer; immunology.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests N.B. serves as an advisor/board member for Apricity, BioNTech, Boehringer Ingelheim, BreakBio, Carisma Therapeutics, CureVac, Genotwin, Gilead, Novartis, PrimeVax, ROME Therapeutics, Tempest Therapeutics, and Rubius Therapeutics and as a consultant for Genentech. N.B. provides research for support for DC Prime, Dragonfly Therapeutics, Inc., Harbor Biomed Sciences, and Regeneron Pharmaceuticals, Inc. N.B. serves on the Scientific Advisory Council/Board of the Cancer Research Institute, Duke University CHAVD, MD Anderson Cancer Center, Parker Institute for Cancer Immunotherapy, and the American Association for Cancer Research. N.B. serves on the grants/research support council for the Cancer Research Institute, Melanoma Research Alliance, Leukemia & Lymphoma Society, Pershing Square Sohn Cancer Research Alliance, and Stand Up to Cancer.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

