Ancient Sheep Genomes Reveal Four Millennia of North European Short-Tailed Sheep in the Baltic Sea Region
- PMID: 38795367
- PMCID: PMC11162877
- DOI: 10.1093/gbe/evae114
Ancient Sheep Genomes Reveal Four Millennia of North European Short-Tailed Sheep in the Baltic Sea Region
Abstract
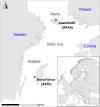
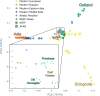
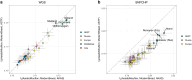
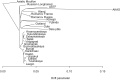
Sheep are among the earliest domesticated livestock species, with a wide variety of breeds present today. However, it remains unclear how far back this diversity goes, with formal documentation only dating back a few centuries. North European short-tailed (NEST) breeds are often assumed to be among the oldest domestic sheep populations, even thought to represent relicts of the earliest sheep expansions during the Neolithic period reaching Scandinavia <6,000 years ago. This study sequenced the genomes (up to 11.6X) of five sheep remains from the Baltic islands of Gotland and Åland, dating from the Late Neolithic (∼4,100 cal BP) to historical times (∼1,600 CE). Our findings indicate that these ancient sheep largely possessed the genetic characteristics of modern NEST breeds, suggesting a substantial degree of long-term continuity of this sheep type in the Baltic Sea region. Despite the wide temporal spread, population genetic analyses show high levels of affinity between the ancient genomes and they also exhibit relatively high genetic diversity when compared to modern NEST breeds, implying a loss of diversity in most breeds during the last centuries associated with breed formation and recent bottlenecks. Our results shed light on the development of breeds in Northern Europe specifically as well as the development of genetic diversity in sheep breeds, and their expansion from the domestication center in general.
Keywords: Baltic; ancient DNA; breeds; domestication; sheep.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2018 [accessed 2022 Apr 12]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/2010.
-
- Apel J, Wallin P, Storå J, Possnert G. Early Holocene human population events on the island of Gotland in the Baltic Sea (9200-3800 cal. BP). Quat Int. 2018:465:276–286. 10.1016/j.quaint.2017.03.044. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

