Lipidome atlas of the adult human brain
- PMID: 38796479
- PMCID: PMC11127996
- DOI: 10.1038/s41467-024-48734-y
Lipidome atlas of the adult human brain
Abstract
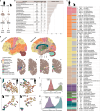
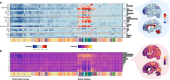
Lipids are the most abundant but poorly explored components of the human brain. Here, we present a lipidome map of the human brain comprising 75 regions, including 52 neocortical ones. The lipidome composition varies greatly among the brain regions, affecting 93% of the 419 analyzed lipids. These differences reflect the brain's structural characteristics, such as myelin content (345 lipids) and cell type composition (353 lipids), but also functional traits: functional connectivity (76 lipids) and information processing hierarchy (60 lipids). Combining lipid composition and mRNA expression data further enhances functional connectivity association. Biochemically, lipids linked with structural and functional brain features display distinct lipid class distribution, unsaturation extent, and prevalence of omega-3 and omega-6 fatty acid residues. We verified our conclusions by parallel analysis of three adult macaque brains, targeted analysis of 216 lipids, mass spectrometry imaging, and lipidome assessment of sorted murine neurons.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

