Oncofetal SNRPE promotes HCC tumorigenesis by regulating the FGFR4 expression through alternative splicing
- PMID: 38796598
- PMCID: PMC11231362
- DOI: 10.1038/s41416-024-02689-5
Oncofetal SNRPE promotes HCC tumorigenesis by regulating the FGFR4 expression through alternative splicing
Abstract
Background: Due to insufficient knowledge about key molecular events, Hepatocellular carcinoma (HCC) lacks effective treatment targets. Spliceosome-related genes were significantly altered in HCC. Oncofetal proteins are ideal tumor therapeutic targets. Screening of differentially expressed Spliceosome-related oncofetal protein in embryonic liver development and HCC helps discover effective therapeutic targets for HCC.
Methods: Differentially expressed spliceosome genes were analysis in fetal liver and HCC through bioinformatics analysis. Small nuclear ribonucleoprotein polypeptide E (SNRPE) expression was detected in fetal liver, adult liver and HCC tissues. The role of SNRPE in HCC was performed multiple assays in vitro and in vivo. SNRPE-regulated alternative splicing was recognized by RNA-Seq and confirmed by multiple assays.
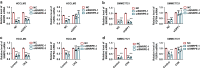
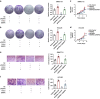
Results: We herein identified SNRPE as a crucial oncofetal splicing factor, significantly associated with the adverse prognosis of HCC. SOX2 was identified as the activator for SNRPE reactivation. Efficient knockdown of SNRPE resulted in the complete cessation of HCC tumorigenesis and progression. Mechanistically, SNRPE knockdown reduced FGFR4 mRNA expression by triggering nonsense-mediated RNA decay. A partial inhibition of SNRPE-induced malignant progression of HCC cells was observed upon FGFR4 knockdown.
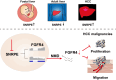
Conclusions: Our findings highlight SNRPE as a novel oncofetal splicing factor and shed light on the intricate relationship between oncofetal splicing factors, splicing events, and carcinogenesis. Consequently, SNRPE emerges as a potential therapeutic target for HCC treatment. Model of oncofetal SNRPE promotes HCC tumorigenesis by regulating the AS of FGFR4 pre-mRNA.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

