Bacterial hosts of clinically significant beta-lactamase genes in Croatian wastewaters
- PMID: 38796694
- PMCID: PMC11165274
- DOI: 10.1093/femsec/fiae081
Bacterial hosts of clinically significant beta-lactamase genes in Croatian wastewaters
Abstract
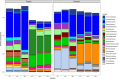
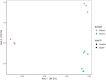
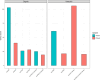
Wastewater treatment plants (WWTPs) provide a suitable environment for the interaction of antibiotic resistant bacteria and antibiotic-resistance genes (ARGs) from human, animal, and environmental sources. The aim was to study the influent and effluent of two WWTPs in Croatia to identify bacterial hosts of clinically important beta-lactamase genes (blaTEM, blaVIM, blaOXA-48-like) and observe how their composition changes during the treatment process. A culture-independent epicPCR (Emulsion, Paired isolation and Concatenation Polymerase Chain Reaction) was used to identify the ARG hosts, and 16S rRNA amplicon sequencing to study the entire bacterial community. Different wastewater sources contributed to the significant differences in bacterial composition of the wastewater between the two WWTPs studied. A total of 167 genera were detected by epicPCR, with the Arcobacter genus, in which all ARGs studied were present, dominating in both WWTPs. In addition, the clinically important genera Acinetobacter and Aeromonas contained all ARGs examined. The blaOXA-48-like gene had the highest number of hosts, followed by blaVIM, while blaTEM had the narrowest host range. Based on 16S rRNA gene sequencing, ARG hosts were detected in both abundant and rare taxa. The number of hosts carrying investigated ARGs was reduced by wastewater treatment. EpicPCR provided valuable insights into the bacterial hosts of horizontally transmissible beta-lactamase genes in Croatian wastewater.
Keywords: 16S rRNA; Croatia; antibiotic-resistance genes; bacteria; epicPCR; wastewater.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures


Similar articles
-
Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent.FEMS Microbiol Ecol. 2018 Apr 1;94(4):fiy038. doi: 10.1093/femsec/fiy038. FEMS Microbiol Ecol. 2018. PMID: 29514229 Free PMC article.
-
Characterisation of microbial communities and quantification of antibiotic resistance genes in Italian wastewater treatment plants using 16S rRNA sequencing and digital PCR.Sci Total Environ. 2024 Jul 10;933:173217. doi: 10.1016/j.scitotenv.2024.173217. Epub 2024 May 13. Sci Total Environ. 2024. PMID: 38750766
-
Sensitivity and consistency of long- and short-read metagenomics and epicPCR for the detection of antibiotic resistance genes and their bacterial hosts in wastewater.J Hazard Mater. 2024 May 5;469:133939. doi: 10.1016/j.jhazmat.2024.133939. Epub 2024 Mar 7. J Hazard Mater. 2024. PMID: 38490149
-
Microbiome and antibiotic resistance profiling in submarine effluent-receiving coastal waters in Croatia.Environ Pollut. 2022 Jan 1;292(Pt A):118282. doi: 10.1016/j.envpol.2021.118282. Epub 2021 Oct 4. Environ Pollut. 2022. PMID: 34619178 Review.
-
Distribution, sources, and potential risks of antibiotic resistance genes in wastewater treatment plant: A review.Environ Pollut. 2022 Oct 1;310:119870. doi: 10.1016/j.envpol.2022.119870. Epub 2022 Jul 31. Environ Pollut. 2022. PMID: 35921944 Review.
References
-
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (27 July 2023, date last accessed).
-
- Bates B, Maechler M, Bolker B et al. Fitting linear mixed-effects models using lme4. J Stat Soft. 2015;67:1–48.

