Neuroglobin overexpression in cerebellar neurons of Harlequin mice improves mitochondrial homeostasis and reduces ataxic behavior
- PMID: 38796706
- PMCID: PMC11286817
- DOI: 10.1016/j.ymthe.2024.05.030
Neuroglobin overexpression in cerebellar neurons of Harlequin mice improves mitochondrial homeostasis and reduces ataxic behavior
Abstract
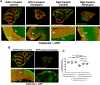
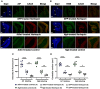
Neuroglobin, a member of the globin superfamily, is abundant in the brain, retina, and cerebellum of mammals and localizes to mitochondria. The protein exhibits neuroprotective capacities by participating in electron transfer, oxygen supply, and protecting against oxidative stress. Our objective was to determine whether neuroglobin overexpression can be used to treat neurological disorders. We chose Harlequin mice, which harbor a retroviral insertion in the first intron of the apoptosis-inducing factor gene resulting in the depletion of the corresponding protein essential for mitochondrial biogenesis. Consequently, Harlequin mice display degeneration of the cerebellum and suffer from progressive blindness and ataxia. Cerebellar ataxia begins in Harlequin mice at the age of 4 months and is characterized by neuronal cell disappearance, bioenergetics failure, and motor and cognitive impairments, which aggravated with aging. Mice aged 2 months received adeno-associated viral vectors harboring the coding sequence of neuroglobin or apoptosis-inducing factor in both cerebellar hemispheres. Six months later, Harlequin mice exhibited substantial improvements in motor and cognitive skills; probably linked to the preservation of respiratory chain function, Purkinje cell numbers and connectivity. Thus, without sharing functional properties with apoptosis-inducing factor, neuroglobin was efficient in reducing ataxia in Harlequin mice.
Keywords: AAV2/9 vectors; Apoptosis-inducing factor; Harlequin mice; Purkinje cells; gene therapy; mitochondria; neuroglobin; respiratory chain.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests An application for a patent has been filed on neuroglobin in Europe (EP22306002.1) and the United States (PCTEP2023/068369) in 2022 and 2023 (M.C.-D. and P.G).
Figures







References
-
- Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T.J., Goodman R.P., Grabarek Z., Haas M.E., Hung W.H.W., et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49:D1541–D1547. doi: 10.1093/nar/gkaa1011. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

