Multi-omic integration reveals alterations in nasal mucosal biology that mediate air pollutant effects on allergic rhinitis
- PMID: 38796780
- PMCID: PMC11560721
- DOI: 10.1111/all.16174
Multi-omic integration reveals alterations in nasal mucosal biology that mediate air pollutant effects on allergic rhinitis
Abstract
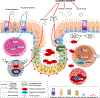
Background: Allergic rhinitis is a common inflammatory condition of the nasal mucosa that imposes a considerable health burden. Air pollution has been observed to increase the risk of developing allergic rhinitis. We addressed the hypotheses that early life exposure to air toxics is associated with developing allergic rhinitis, and that these effects are mediated by DNA methylation and gene expression in the nasal mucosa.
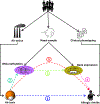
Methods: In a case-control cohort of 505 participants, we geocoded participants' early life exposure to air toxics using data from the US Environmental Protection Agency, assessed physician diagnosis of allergic rhinitis by questionnaire, and collected nasal brushings for whole-genome DNA methylation and transcriptome profiling. We then performed a series of analyses including differential expression, Mendelian randomization, and causal mediation analyses to characterize relationships between early life air toxics, nasal DNA methylation, nasal gene expression, and allergic rhinitis.
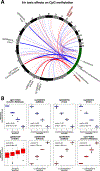
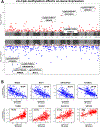
Results: Among the 505 participants, 275 had allergic rhinitis. The mean age of the participants was 16.4 years (standard deviation = 9.5 years). Early life exposure to air toxics such as acrylic acid, phosphine, antimony compounds, and benzyl chloride was associated with developing allergic rhinitis. These air toxics exerted their effects by altering the nasal DNA methylation and nasal gene expression levels of genes involved in respiratory ciliary function, mast cell activation, pro-inflammatory TGF-β1 signaling, and the regulation of myeloid immune cell function.
Conclusions: Our results expand the range of air pollutants implicated in allergic rhinitis and shed light on their underlying biological mechanisms in nasal mucosa.
Keywords: DNA methylation; air pollution; allergic rhinitis; allergy; causal mediation; epigenome; gene expression; multi‐omic; nasal; systems biology; transcriptome.
© 2024 European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
Conflict of interest
The authors have declared that no conflict of interest exists.
Figures


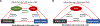
References
-
- Zhang Y, Lan F, Zhang L. Advances and highlights in allergic rhinitis. Allergy 2021; 76:3383–9. - PubMed
-
- Dierick BJH, van der Molen T, Flokstra-de Blok BMJ, Muraro A, Postma MJ, Kocks JWH, van Boven JFM. Burden and socioeconomics of asthma, allergic rhinitis, atopic dermatitis and food allergy. Expert Rev Pharmacoecon Outcomes Res 2020; 20:437–53. - PubMed
-
- Vandenplas O, Vinnikov D, Blanc PD, Agache I, Bachert C, Bewick M, et al. Impact of Rhinitis on Work Productivity: A Systematic Review. Journal of Allergy and Clinical Immunology-in Practice 2018; 6:1274–+. - PubMed
-
- Devillier P, Bousquet J, Salvator H, Naline E, Grassin-Delyle S, de Beaumont O. In allergic rhinitis, work, classroom and activity impairments are weakly related to other outcome measures. Clinical and Experimental Allergy 2016; 46:1456–64. - PubMed
-
- Colas C, Brosa M, Anton E, Montoro J, Navarro A, Dordal MT, et al. Estimate of the total costs of allergic rhinitis in specialized care based on real-world data: the FERIN Study. Allergy 2017; 72:959–66. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

