This is a preprint.
IFIT1 is rapidly evolving and exhibits disparate antiviral activities across 11 mammalian orders
- PMID: 38798375
- PMCID: PMC11118533
- DOI: 10.1101/2024.05.13.593954
IFIT1 is rapidly evolving and exhibits disparate antiviral activities across 11 mammalian orders
Update in
-
IFIT1 is rapidly evolving and exhibits disparate antiviral activities across 11 mammalian orders.Elife. 2025 Oct 22;13:RP101929. doi: 10.7554/eLife.101929. Elife. 2025. PMID: 41123582 Free PMC article.
Abstract
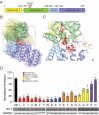
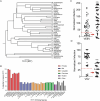
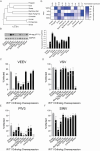
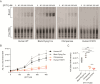
Mammalian mRNAs possess an N7-methylguanosine (m7G) cap and 2'O methylation of the initiating nucleotide at their 5' end, whereas certain viral RNAs lack these characteristic features. The human antiviral restriction factor IFIT1 recognizes and binds to specific viral RNAs that lack the 5' features of host mRNAs, resulting in targeted suppression of viral RNA translation. This interaction imposes significant host-driven evolutionary pressures on viruses, and many viruses have evolved mechanisms to evade the antiviral action of human IFIT1. However, little is known about the virus-driven pressures that may have shaped the antiviral activity of IFIT1 genes across mammals. Here, we take an evolution-guided approach to show that the IFIT1 gene is rapidly evolving in multiple mammalian clades, with positive selection acting upon several residues in distinct regions of the protein. In functional assays with 39 IFIT1s spanning diverse mammals, we demonstrate that IFIT1 exhibits a range of antiviral phenotypes, with many orthologs lacking antiviral activity against viruses that are strongly suppressed by other IFIT1s. We further show that IFIT1s from human and a bat, the black flying fox, inhibit Venezuelan equine encephalitis virus (VEEV) and strongly bind to Cap0 RNAs. Unexpectedly, chimpanzee IFIT1, which differs from human IFIT1 by only 8 amino acids, does not inhibit VEEV infection and exhibits minimal Cap0 RNA-binding. In mutagenesis studies, we determine that amino acids 364 and 366, the latter of which is rapidly evolving, are sufficient to confer the differential anti-VEEV activity between human and chimpanzee IFIT1. These data suggest that virus-host genetic conflicts have influenced the antiviral specificity of IFIT1 across diverse mammalian orders.
Figures

References
-
- Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, et al. IFIT1 is an antiviral protein that recognizes 5’-triphosphate RNA. Nat Immunol. 2011;12(7):624–30. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
