This is a preprint.
TOR signaling regulates GPCR levels on the plasma membrane and suppresses the Saccharomyces cerevisiae mating pathway
- PMID: 38798445
- PMCID: PMC11118302
- DOI: 10.1101/2024.05.09.593412
TOR signaling regulates GPCR levels on the plasma membrane and suppresses the Saccharomyces cerevisiae mating pathway
Update in
-
TOR signaling regulates GPCR levels on the plasma membrane and suppresses the Saccharomyces cerevisiae mating pathway.J Biol Chem. 2025 Oct;301(10):110700. doi: 10.1016/j.jbc.2025.110700. Epub 2025 Sep 11. J Biol Chem. 2025. PMID: 40945725 Free PMC article.
Abstract
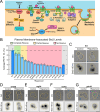
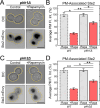
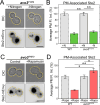
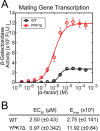
Target of Rapamycin (TOR) complexes and G-protein-coupled receptors (GPCRs) are crucial signaling hubs that coordinate adaptive responses to environmental inputs. While GPCR-mediated regulation of TOR has been extensively studied, little is known about TOR-mediated regulation of GPCRs. Here, we establish TOR as a regulator of GPCR signaling via its control of receptor endocytosis in the yeast mating system. By pairing fluorescence microscopy with yeast genetic approaches, we identify the machinery that bridges TOR nutrient sensing to GPCR internalization. Furthermore, we find that autophagic machinery is employed during mating to deliver active receptor to the vacuole (lysosome), suppressing the mating pathway. These results demonstrate that TOR regulates the localization and signaling of the yeast mating GPCR in both ligand-dependent and -independent contexts. These pathways are highly conserved, suggesting that TOR-regulation of GPCRs may be a broadly conserved mechanism for integrating competing signals involving metabolic state and external communications.
Conflict of interest statement
Competing interests: Authors declare that they have no competing interests.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
