This is a preprint.
Acceleration of genome rearrangement in clitellate annelids
- PMID: 38798472
- PMCID: PMC11118384
- DOI: 10.1101/2024.05.12.593736
Acceleration of genome rearrangement in clitellate annelids
Abstract
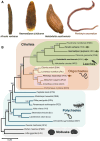
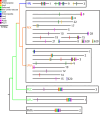
Comparisons of multiple metazoan genomes have revealed the existence of ancestral linkage groups (ALGs), genomic scaffolds sharing sets of orthologous genes that have been inherited from ancestral animals for hundreds of millions of years (Simakov et al. 2022; Schultz et al. 2023) These ALGs have persisted across major animal taxa including Cnidaria, Deuterostomia, Ecdysozoa and Spiralia. Notwithstanding this general trend of chromosome-scale conservation, ALGs have been obliterated by extensive genome rearrangements in certain groups, most notably including Clitellata (oligochaetes and leeches), a group of easily overlooked invertebrates that is of tremendous ecological, agricultural and economic importance (Charles 2019; Barrett 2016). To further investigate these rearrangements, we have undertaken a comparison of 12 clitellate genomes (including four newly sequenced species) and 11 outgroup representatives. We show that these rearrangements began at the base of the Clitellata (rather than progressing gradually throughout polychaete annelids), that the inter-chromosomal rearrangements continue in several clitellate lineages and that these events have substantially shaped the evolution of the otherwise highly conserved Hox cluster.
Keywords: Annelida; Chromosome; Clitellata; Genome; Genome Evolution; Hox Genes; Synteny.
Conflict of interest statement
ETHICS DECLARATIONS All authors declare that they have no competing interests.
Figures

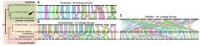
References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J.. 1990. “Basic Local Alignment Search Tool.” Journal of Molecular Biology 215 (3): 403–10. - PubMed
-
- Babenko Vladislav V., Podgorny Oleg V., Manuvera Valentin A., Kasianov Artem S., Manolov Alexander I., Grafskaia Ekaterina N., Shirokov Dmitriy A., et al. 2020. “Draft Genome Sequences of Hirudo Medicinalis and Salivary Transcriptome of Three Closely Related Medicinal Leeches.” BMC Genomics 21 (1): 331. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
