Structure-Based Discovery of Allosteric Inhibitors Targeting a New Druggable Site in the Respiratory Syncytial Virus Polymerase
- PMID: 38799318
- PMCID: PMC11112712
- DOI: 10.1021/acsomega.4c01207
Structure-Based Discovery of Allosteric Inhibitors Targeting a New Druggable Site in the Respiratory Syncytial Virus Polymerase
Abstract
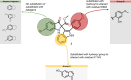
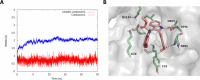
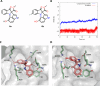
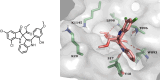
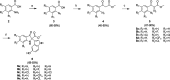
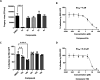
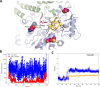
Respiratory syncytial virus (RSV) is a major cause of severe lower respiratory infections for which effective treatment options remain limited. Herein, we employed a computational structure-based design strategy aimed at identifying potential targets for a new class of allosteric inhibitors. Our investigation led to the discovery of a previously undisclosed allosteric binding site within the RSV polymerase, the large (L) protein. This discovery was achieved through a combination of virtual screening and molecular dynamics simulations. Subsequently, we identified two inhibitors, 6a and 10b, which both exhibited promising antiviral activity in the low micromolar range. Resistance profiling revealed a distinctive pattern in how RSV evaded treatment with this class of inhibitors. This pattern strongly suggested that this class of small molecules was targeting a new binding site in the RSV L protein, aligning with the computational predictions made in our study. This study paves the way for the development of more potent inhibitors for combating RSV infections by targeting a new druggable pocket within the RdRp which does not overlap with previously known resistance sites.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): The authors declare the following conflict of interest. A.K.O, L.B., F.G.W, and D.J.M. are inventors on U.S. patent application no. (63/631,132) describing the compounds and their method of use.
Figures




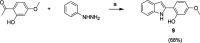
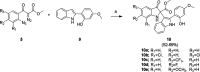

References
LinkOut - more resources
Full Text Sources
Research Materials
