Hepatitis B-related hepatocellular carcinoma: classification and prognostic model based on programmed cell death genes
- PMID: 38799437
- PMCID: PMC11116790
- DOI: 10.3389/fimmu.2024.1411161
Hepatitis B-related hepatocellular carcinoma: classification and prognostic model based on programmed cell death genes
Abstract
Instruction: Hepatitis B virus (HBV) infection is a major risk factor for hepatocellular carcinoma (HCC). Programmed cell death (PCD) is a critical process in suppressing tumor growth, and alterations in PCD-related genes may contribute to the progression of HBV-HCC. This study aims to develop a prognostic model that incorporates genomic and clinical information based on PCD-related genes, providing novel insights into the molecular heterogeneity of HBV-HCC through bioinformatics analysis and experimental validation.
Methods: In this study, we analyzed 139 HBV-HCC samples from The Cancer Genome Atlas (TCGA) and validated them with 30 samples from the Gene Expression Omnibus (GEO) database. Various bioinformatics tools, including differential expression analysis, gene set variation analysis, and machine learning algorithms were used for comprehensive analysis of RNA sequencing data from HBV-HCC patients. Furthermore, among the PCD-related genes, we ultimately chose DLAT for further research on tissue chips and patient cohorts. Besides, immunohistochemistry, qRT-PCR and Western blot analysis were conducted.
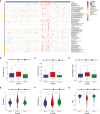
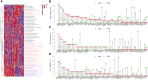
Results: The cluster analysis identified three distinct subgroups of HBV-HCC patients. Among them, Cluster 2 demonstrated significant activation in DNA replication-related pathways and tumor-related processes. Analysis of copy number variations (CNVs) of PCD-related genes also revealed distinct patterns in the three subgroups, which may be associated with differences in pathway activation and survival outcomes. DLAT in tumor tissues of HBV-HCC patients is upregulated.
Discussion: Based on the PCD-related genes, we developed a prognostic model that incorporates genomic and clinical information and provided novel insights into the molecular heterogeneity of HBV-HCC. In our study, we emphasized the significance of PCD-related genes, particularly DLAT, which was examined in vitro to explore its potential clinical implications.
Keywords: clinical characteristics; hepatitis B virus infection; hepatocellular carcinoma; prognostic model; programmed cell death.
Copyright © 2024 Tian, Meng, Yang, Song, Jiang and Zou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Ndow G, Vo-Quang E, Shimakawa Y, Ceesay A, Tamba S, Njai HF, et al. . Clinical characteristics and outcomes of patients with cirrhosis and hepatocellular carcinoma in The Gambia, west Africa: a prospective cohort study. Lancet Glob Health. (2023) 11:e1383–92. doi: 10.1016/S2214-109X(23)00263-2 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

