Ablation of Atp5if1 impairs metabolic reprogramming and proliferation of T lymphocytes and compromises mouse survival
- PMID: 38799559
- PMCID: PMC11126974
- DOI: 10.1016/j.isci.2024.109863
Ablation of Atp5if1 impairs metabolic reprogramming and proliferation of T lymphocytes and compromises mouse survival
Abstract
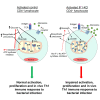
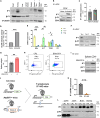
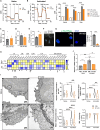
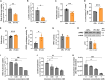
T cells experience metabolic reprogramming to an enhanced glycolysis upon activation. Herein, we have investigated whether ATPase Inhibitory Factor 1 (IF1), the physiological inhibitor of mitochondrial ATP synthase, participates in rewiring T cells to a particular metabolic phenotype. We show that the activation of naive CD4+ T lymphocytes both in vitro and in vivo is accompanied by a sharp upregulation of IF1, which is expressed only in Th1 effector cells. T lymphocytes of conditional CD4+-IF1-knockout mice display impaired glucose uptake and flux through glycolysis, reducing the biogenesis of mitochondria and cellular proliferation after activation. Consequently, mice devoid of IF1 in T lymphocytes cannot mount an effective Th1 response against bacterial infection compromising their survival. Overall, we show that the inhibition of a fraction of ATP synthase by IF1 regulates metabolic reprogramming and functionality of T cells, highlighting the essential role of IF1 in adaptive immune responses.
Keywords: Biological sciences; Immunology; Molecular biology; Physiology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
IF1 ablation prevents ATP synthase oligomerization, enhances mitochondrial ATP turnover and promotes an adenosine-mediated pro-inflammatory phenotype.Cell Death Dis. 2023 Jul 12;14(7):413. doi: 10.1038/s41419-023-05957-z. Cell Death Dis. 2023. PMID: 37433784 Free PMC article.
-
Regulation of the H+-ATP synthase by IF1: a role in mitohormesis.Cell Mol Life Sci. 2017 Jun;74(12):2151-2166. doi: 10.1007/s00018-017-2462-8. Epub 2017 Feb 6. Cell Mol Life Sci. 2017. PMID: 28168445 Free PMC article. Review.
-
A Review of the Inhibition of the Mitochondrial ATP Synthase by IF1 in vivo: Reprogramming Energy Metabolism and Inducing Mitohormesis.Front Physiol. 2018 Sep 19;9:1322. doi: 10.3389/fphys.2018.01322. eCollection 2018. Front Physiol. 2018. PMID: 30283362 Free PMC article. Review.
-
The ATPase Inhibitory Factor 1 (IF1) Contributes to the Warburg Effect and Is Regulated by Its Phosphorylation in S39 by a Protein Kinase A-like Activity.Cancers (Basel). 2024 Feb 29;16(5):1014. doi: 10.3390/cancers16051014. Cancers (Basel). 2024. PMID: 38473373 Free PMC article. Review.
-
F-ATP synthase inhibitory factor 1 regulates metabolic reprogramming involving its interaction with c-Myc and PGC1α.Front Oncol. 2023 Jul 3;13:1207603. doi: 10.3389/fonc.2023.1207603. eCollection 2023. Front Oncol. 2023. PMID: 37469400 Free PMC article.
Cited by
-
Oxidative Phosphorylation in Uncoupled Mitochondria.Bioessays. 2025 Sep;47(9):e70038. doi: 10.1002/bies.70038. Epub 2025 Jul 6. Bioessays. 2025. PMID: 40619853 Free PMC article. Review.
References
-
- Boyer P.D. The ATP synthase. A splendid molecular machine. Annu. Rev. Biochem. 1997;66:717–749. - PubMed
-
- Walker J.E. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013;41:1–16. - PubMed
-
- Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M.R., Abramov A.Y., Tinker A., Duchen M.R. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. - PubMed
-
- Kuhlbrandt W. Structure and Mechanisms of F-Type ATP Synthases. Annu. Rev. Biochem. 2019;88:515–549. - PubMed
-
- Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

