Investigating and Quantifying Molecular Complexity Using Assembly Theory and Spectroscopy
- PMID: 38799656
- PMCID: PMC11117308
- DOI: 10.1021/acscentsci.4c00120
Investigating and Quantifying Molecular Complexity Using Assembly Theory and Spectroscopy
Abstract
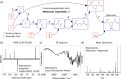
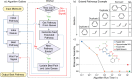
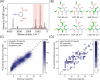
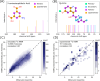
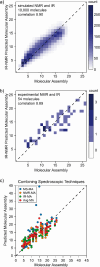
Current approaches to evaluate molecular complexity use algorithmic complexity, rooted in computer science, and thus are not experimentally measurable. Directly evaluating molecular complexity could be used to study directed vs undirected processes in the creation of molecules, with potential applications in drug discovery, the origin of life, and artificial life. Assembly theory has been developed to quantify the complexity of a molecule by finding the shortest path to construct the molecule from building blocks, revealing its molecular assembly index (MA). In this study, we present an approach to rapidly infer the MA of molecules from spectroscopic measurements. We demonstrate that the MA can be experimentally measured by using three independent techniques: nuclear magnetic resonance (NMR), tandem mass spectrometry (MS/MS), and infrared spectroscopy (IR). By identifying and analyzing the number of absorbances in IR spectra, carbon resonances in NMR, or molecular fragments in tandem MS, the MA of an unknown molecule can be reliably estimated. This represents the first experimentally quantifiable approach to determining molecular assembly. This paves the way to use experimental techniques to explore the evolution of complex molecules as well as a unique marker of where an evolutionary process has been operating.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Adams K.; Coley C. W.. Equivariant Shape-Conditioned Generation of 3D Molecules for Ligand-Based Drug Design. 2022,10.48550/ARXIV.2210.04893. - DOI
LinkOut - more resources
Full Text Sources
