Translating nanoEHS data using EPA NaKnowBase and the resource description framework
- PMID: 38800073
- PMCID: PMC11128042
- DOI: 10.12688/f1000research.141056.1
Translating nanoEHS data using EPA NaKnowBase and the resource description framework
Abstract
Background: The U.S. Federal Government has supported the generation of extensive amounts of nanomaterials and related nano Environmental Health and Safety (nanoEHS) data, there is a need to make these data available to stakeholders. With recent efforts, a need for improved interoperability, translation, and sustainability of Federal nanoEHS data in the United States has been realized. The NaKnowBase (NKB) is a relational database containing experimental results generated by the EPA Office of Research and Development (ORD) regarding the actions of engineered nanomaterials on environmental and biological systems. Through the interaction of the National Nanotechnology Initiative's Nanotechnology Environmental Health Implications (NEHI) Working Group, and the Database and Informatics Interest Group (DIIG), a U.S. Federal nanoEHS Consortium has been formed.
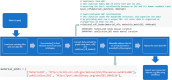
Methods: The primary goal of this consortium is to establish a "common language" for nanoEHS data that aligns with FAIR data standards. A second goal is to overcome nomenclature issues inherent to nanomaterials data, ultimately allowing data sharing and interoperability across the diverse U.S. Federal nanoEHS data compendium, but also in keeping a level of consistency that will allow interoperability with U.S. and European partners. The most recent version of the EPA NaKnowBase (NKB) has been implemented for semantic integration. Computational code has been developed to use each NKB record as input, modify and filter table data, and subsequently output each modified record to a Research Description Framework (RDF). To improve the accuracy and efficiency of this process the EPA has created the OntoSearcher tool. This tool partially automates the ontology mapping process, thereby reducing onerous manual curation.
Conclusions: Here we describe the efforts of the US EPA in promoting FAIR data standards for Federal nanoEHS data through semantic integration, as well as in the development of NAMs (computational tools) to facilitate these improvements for nanoEHS data at the Federal partner level.
Keywords: nanomaterial; database; ontology.
Copyright: © 2024 Mortensen HM et al.
Conflict of interest statement
No competing interests were disclosed.
Figures






References
Bibliography
-
- Allied Market Research: Nanotechnology Market By Type (Nanosensor and Nanodevice) and Application (Electronics, Energy, Chemical Manufacturing, Aerospace & Defense, Healthcare, and Others): Global Opportunity Analysis and Industry Forecast, 2021-2030. 2021. Reference Source
-
- Ayadi A, Auffan M, Rose J: Ontology-based NLP information extraction to enrich nanomaterial environmental exposure database. Procedia Comput. Sci. 2020;176:360–369. 10.1016/j.procs.2020.08.037 - DOI
-
- Bachmann M: RapidFuzz. Zenodo. 2021.
-
- Boettiger C: rdflib: A high level wrapper around the redland package for common rdf applications. Zenodo. 2018.
References
-
- : Harmonising knowledge for safer materials via the “NanoCommons” Knowledge Base. Frontiers in Physics .2023;11: 10.3389/fphy.2023.1271842 10.3389/fphy.2023.1271842 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

